DeepLung: A Novel Lung Cancer Recurrence Prediction Model Using Deep Learning.
Kairui Yang
The Stony Brook School, 1 Chapman Pkwy, Stony Brook, NY 11790, USA
*Corresponding Author: Kairui Yang, The Stony Brook School, 1 Chapman Pkwy, Stony Brook, NY 11790, USA
Received: 16 August 2025; Accepted: 25 August 2025; Published: 05 September 2025.
Article Information
Citation: Kairui Yang. Rutin: DeepLung: A Novel Lung Cancer Recurrence Prediction Model Using Deep Learning. Journal of Bioinformatics and Systems Biology. 8 (2025): 58-67.
View / Download Pdf Share at FacebookAbstract
Lung cancer recurrence represents a critical determinant of patient prognosis, posing a significant threat to survival outcomes. The development of reliable recurrence prediction tools is therefore clinically imperative to guide therapeutic decision-making and improve both survival and quality of life. The model comprehensively analyzes tissue features from all designated regions of interest (ROIs) identified in pathological reports to predict lung cancer relapse probability.Validation through timedependent receiver operating characteristic (ROC) analysis demonstrated robust predictive performance. Survival analysis using semi-parametric Cox proportional hazards models confirmed the model's superiority over conventional TNM staging, with statistically significant improvements in AUC values (p<0.05). This prediction model exhibits substantial clinical translational potential, providing a valuable foundation for personalized treatment strategies and emerging as a novel decision-support tool for prognostic management.
Keywords
<p>Deep Learning, Lung Cancer Recurrence, Survival Analysis, CT, HE.</p>
Article Details
1. Introduction
Lung cancer has always been a dilemma for scientists, doctors, patients and caregivers [1]. There were 238,340 new lung cancer cases in the US [2] and 127,070 deaths from it estimated in 2023. One of the more alarming aspects of lung cancer is its abnormally high rates of recurrence [3]. Approximately 39% [4] patients experience recurrence of lung cancer, and roughly 50–90% of postoperative recurrences of lung cancer can happen within 2 years [5-7]. In addition, patients with recurrence benefit less from chemotherapy or targeted therapy [8-10]. Thus for higher survival rates patients tend to undergo local therapies [11] including radiation which leads to damage in lung tissues. Importantly, recurrent disease is associated with significantly poorer outcomes [12], demonstrating a median survival of just 31 months compared to 63.1 months for non-recurrent cases [13]. These clinical realities underscore the critical need for early recurrence detection, which could substantially improve treatment efficacy and overall survival outcomes.
Recent technological advancements [14] have facilitated the development of various predictive models for lung cancer recurrence, offering potential benefits for both patient outcomes and healthcare resource utilization. Current approaches include molecular methods such as ctDNA analysis [15], whole genome sequencing (WGS) [16], and transcriptome panels [17]. While promising, these techniques face clinical implementation challenges due to their substantial costs and restricted dataset availability.
In contrast, imaging-based approaches - particularly radiomic feature extraction from computed tomography (CT) scans [18-19] and hematoxylin & eosin (H&E) stained images [20] - have demonstrated both wider clinical applicability and superior predictive performance. Deep learning applications in medical imaging have shown particular promise in this domain. For instance, Lee et al. [21] developed a CT-based radiomic model for 2-year non-small cell lung cancer (NSCLC) recurrence prediction, achieving 71.42% accuracy, 80.95% sensitivity, 61.90% specificity, and an AUC of 0.74. Similarly, Aopong et al. [22] employed ResNet50 and DenseNet121 architectures for CT-based NSCLC recurrence prediction [23], obtaining AUC scores of 0.6714 and 0.6712 respectively, while potentially reducing reliance on invasive biopsies.
H&E image analysis has also shown potential, with Wang, X [24] reporting 81% training accuracy (82% and 75% in two validation cohorts, separately) through nuclear feature extraction. However, this approach was limited by its focus on small tumor portions and early-stage images, potentially compromising its ability to capture tumor heterogeneity. Importantly, most existing studies share common limitations including single-institution datasets, unoptimized feature selection, and inadequate region of interest (ROI) analysis.
Based on this, our study leverages the complementary strengths of H&E histopathology and CT imaging to achieve multidimensional tumor characterization through Integrated Deep Learning Engine (IDLE) [25], which synergistically combines: (1) pathologist-annotated regions of interest (ROIs) serving as the foundational input material, (2) CT-derived anatomical features to emphasize lung lesions and tumor size and (3) H&E-extracted morphological and biological characteristics including tumor aggressiveness markers and radiographically occult biomarker expression patterns. The fusion of radiomic and histopathologic data significantly enhances model performance by concurrently capturing macroscopic anatomical changes and microscopic tumor behavior. Cox proportional hazards analysis demonstrates IDLE's dual advantage as both an independent prognostic factor (p<0.01) and a superior predictor compared to conventional TNM staging (HR 1.50 for stage II, 2.85 for stage III, and 8.88 for stage IV, P < .001) [26]. Our integrated CT-H&E framework provides crucial clinical benefits: enhanced recurrence prediction through optimized performance metrics, allowing for earlier intervention windows and enabling precision medicine approaches.
2. Material and methods
2.1 Data Screening and Downloading
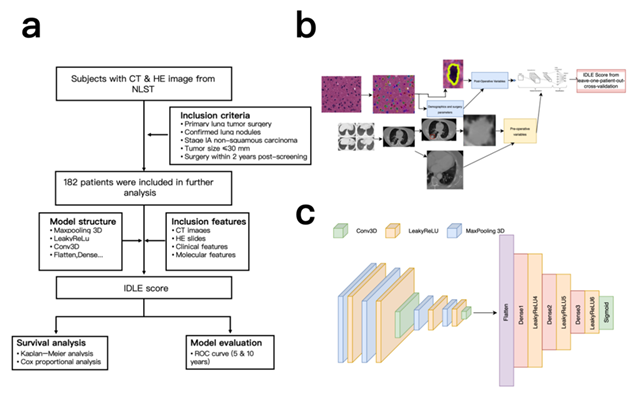
Through material transfer agreements with XLab, the NLST (National Lung Screening Trial) (https://www.cancer.gov/types/lung/research/nlst) provided us with access to a comprehensive set of data, including preoperative LDCT (low-dose computed tomography) images, H&E-stained tissue sections, pathologist reports, and postoperative follow-up information. The study cohort was selected based on the following criteria: (1) patients with a primary lung tumor surgery completion; (2) those with a documented pathologist adjudication of lung nodules in NLST; (3) IA-pathological stage non-squamous lung carcinoma (these individuals are staged through the newest AJCC system) [27]; (4) invasive tumor size up to 30mm; and (5) surgeries conducted within 2 years of completing the final round of screening. Among all, 182 cases met the criteria previously outlined and were included for downstream analysis.
2.2 Image Processing
Through OpenCV [28], the raw CT images were preprocessed by resampling to a fixed voxel spacing, and a specified window width and level were applied for grayscale transformation. Cross-referenced [29] regions of interest (ROIs) of the tumor were utilized, following a methodology outlined in the prior study by Huang et al. [30]. For this method, The weighted center for each voxel was first determined within the segmented lesion.
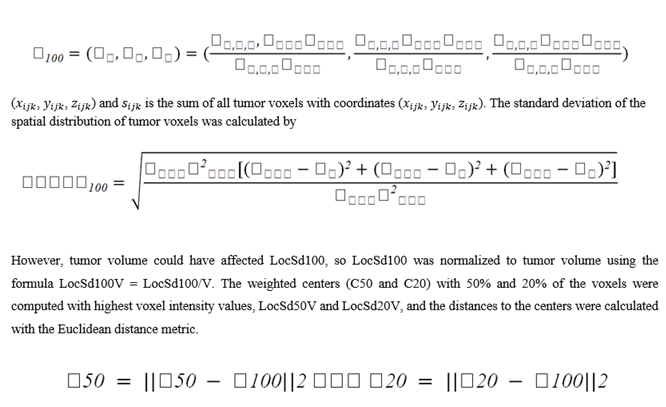
Aerts et al.’s [31] formulas were used to extract the 3D radiomics features. The energy, root mean square (RMS), entropy, and uniformity metrics were computed with a consistent bin width of 100 Hounsfield units. For the second-order gray-level co-occurrence matrices (GLCMs), calculations were performed at angles of 0, 45, 90, 135 degrees in the sagittal, transverse, and coronal orientations, resulting in 13 total directional GLCMs:
(1,0,0); (0,1,0); (1,1,0); (−1,1,0); (1,0,1); (1,1,1); (1,−1,1); (0,0,1); (0,1,1); (0,−1,1); (−1,0,1); (−1,1,1); (−1,−1,1).
22 texture attributes derived from each GLCM, which was then averaged across all 13 orientations to produce the 3D texture features. To extract second-order grayscale run length matrix (GLRLM) features, voxel distances were set at values of d = 2, 3, 4, 5, and 13. From each GLRLM, 11 texture attributes were calculated, thus all 52 orientation-distance combinations were averaged to form the final 3D texture metric.
Additionally, a mean intensity ratio was obtained by dividing the average voxel intensity within the tumor by that of the peritumoral region, providing a comparative measure of tumor versus peritumoral intensity. When calculating the quantile ratio R(q) (qth quantile of voxel intensity within the tumor divided by the qth quantile of voxel intensity in the surrounding peritumoral area) at q = 50 and 90, the quotient of tumor intensity divided by peritumoral intensity for those quantiles yielded a set of 173 features per LDCT image [32].
2.3 Clinical and Molecular Variable Processing
In addition to imaging data, other clinical information was also included to calculate the final IDLE score. Combining patient demographics at the time of surgery, surgical procedure type, residual disease status, whether lymph node dissection was performed, and tissue characteristics extracted from pathology slides and surgical records, each of these variables was coded numerically, as categorical fields are turned into 0/1, and how time intervals or continuous measures are handled as direct numeric columns (provided as supplementary table or figure). This combination of binary, factor, and numeric variables forms the unified input for survival modeling and for computing the IDLE score. The genomics data, comprising expression levels of 5269 selected genes downloaded from NLST, was retrieved using a custom DataLoader tailored for 5-fold cross-validation. Preprocessing steps included normalization to standardize gene expression values across samples, filtering to retain the most informative genes based on variance and biological relevance, and imputation of missing values to maintain data integrity. These preprocessed genomic features were subsequently integrated with PyRadiomics and DenseNet-derived features through concatenation, enabling the construction of multimodal models to predict patient outcomes. This comprehensive preprocessing ensured compatibility across data modalities and enhanced the predictive performance of the models. All of these data points - both clinical (demographic and surgical) and imaging (tumor or peritumoral features)- are merged into a unified feature set.
2.4 Model Construction
Similar to prior publication [33-34], MLP architecture [35] featuring two hidden layers and a final output layer were constructed upon Pytorch [36]. The first hidden layer contains an activation function LeakyReLu based on input data that contains preoperative LDCT lung image characteristics across different anatomical regions as input. Based on the output of the first layer determines the activation function of the subsequent hidden layer LeakyReLU, adjusted by their corresponding weights. Both layers underwent feature selection and weight refinement, utilizing a cross entropy loss function with an L2 regularization term to penalize complexity. The second hidden layer, that is, the input variables and weights were finally passed to a random survival forest, which was the final hidden layer. Finally, random survival forest predicted the value of the network in the range of [0, 1]. The IDLE score was calculated using the leave-one-patient-out cross-validation technique [34].
2.5 Model Evaluation
Demographic and clinical traits of patients who experienced progression and those who did not were then analyzed using summary statistics. Patients were divided into high-risk and low-risk subgroups through the median values of IDLE scores. By calculating the 5-year and 10-year time-dependent area under the ROC curve (AUC), along with time-dependent positive predictive value (PPV), time-dependent negative predictive value (NPV), and the hazard ratio (HR), the accuracy is evaluated to compare progression-free survival between the high-risk and low-risk groups. To assess the precision of the AUC, 500 bootstrap simulations were performed, calculating the standard deviation of the AUC using inverse probability of censoring weighted estimators. This approach allows for robust estimation of model performance, accounting for censoring and providing a more accurate reflection of the model's predictive power.
2.6 Survival Analysis
To evaluate the added benefit of incorporating IDLE scores alongside TNM staging and tumor grade, a multivariate Cox proportional hazards model was used, in which contained six modalities: Genomics (G), Pyradiomics (P), DenseNet (D), early fusion (EF), intermediate fusion (IF), and late fusion (LF). The Cox proportional hazards model [36] built upon survival stands as a prominent semi-parametric approach in survival analysis, and it was used to evaluate the hazard ratio(HR) for each parameter. A log-rank test was conducted to compare Kaplan–Meier survival curves in subgroups with high or low survival rates, utilizing survival (v3.2-3) [37] for the underlying survival analysis framework. The implementation of the algorithm was used from the scikit-survival [38] package.
2.7 Statistical Methods
Continuous variables were presented as median and interquartile range (IQR). We used the Kolmogorov–Smirnov test to assess whether data followed a normal distribution. Depending on the distribution and sample variance, a two-sample t-test, Welch’s two-sample t-test, or Mann–Whitney U test was employed to compare differences in continuous data. Categorical variables were expressed as n%, and a chi-square test or Fisher’s exact test (as appropriate) was used to compare group differences. A p-value less than 0.05 was considered statistically significant. All statistical analyses and figure generation were performed in R (version 3.6.1) [39].
3. Results:
3.1 linical characteristics of included patients from NLST
First, datasets of patients with CT and HE images are downloaded from NLST, selected by the following criteria. 182 patients were ultimately included and used for the model. (Figure 1a)
Table 1 outlines the demographic and clinical profiles of the 182 selected patients. Over a 12-year follow-up period, 54 individuals experienced cancer progression. There were no significant differences in age at surgery, smoking history, tumor location, surgical procedure, size of the excised lesion, or largest invasive tumor size between patients with and without cancer progression (all p > 0.10, Table 1). However, the incidence of recurrence was significantly delayed in patients who experienced progression (39%, 21/54) compared to those without progression (21%, 27/128, p = 0.0167, Table 1) in a way that malignancies were detected more than six months after the final low-dose CT (LDCT) screening.
The interval between the last preoperative LDCT and surgery was longer in patients with cancer progression (p = 0.0468). All patients initially received surgical treatment, except for three who underwent chemotherapy or radiotherapy two to three months prior to surgery. Among the patients, 84% underwent lobectomy, and 97% had no residual disease post-surgery. Additionally, 90% (164/182) had negative lymphadenectomy results.
Table1: A summary of the participant group. The advancement of cancer is characterized by any of the following occurrences within a 12-year observation window post-surgical removal of the primary tumor: the return of lung cancer, the spread of lung cancer to other parts of the body, or death resulting from lung cancer.
|
No Progression N = 128 |
No Progression N = 54 |
p1 |
||
|
Cancers diagnosed 6 months after the last LDCT screening date |
27 |
21 |
0.0167 |
|
|
Lung cancer-related death |
0 |
45 |
||
|
Age at surgery |
64.74.9 |
65.94.8 |
0.1067 |
|
|
Female,N(%) |
58(45%) |
24(44%) |
1 |
|
|
Smoke pack-years |
6629 |
7241 |
0.2889 |
|
|
Days from the last LDCT screening to the date of lung surgery |
177210 |
267299 |
0.0468 |
|
|
surgery type |
Sublobar resection Lobe ctomy |
21 |
8 |
1 |
|
107 |
46 |
|||
|
Lymphadenectomy |
N(%) |
115(90%) |
49(91%) |
1 |
|
Residual disease after surgery |
R0 |
124 |
53 |
1 |
|
R1 |
4 |
1 |
||
|
Surgically removed lesion size(mm) |
1.28691 |
1.2584 |
0.6713 |
|
|
Largest invasive tumor size(mm) |
11.46.08 |
0.56671 |
0.1604 |
|
|
Pathological cancer stage(TNM, 8th edition) |
IA1(T1a) |
50 |
19 |
|
|
IA1(T1b) |
63 |
30 |
1 |
|
|
IA1(T1c) |
15 |
5 |
||
|
Highest tumor grade from all the ROIS |
1=well-differentiated |
34 |
4 |
|
|
2=moderately differentiated |
||||
|
3=poorly differentiated |
53 |
30 |
||
|
4=undifferentiated |
||||
|
Undetermined(GX) |
35 |
14 |
0.0163 |
|
|
5 |
4 |
|||
|
1 |
2 |
3.2 Fold-wise variability and modality-fusion effects
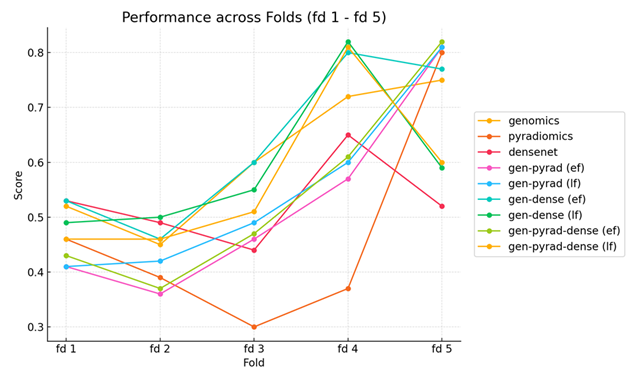
To systematically assess the relative contribution of distinct feature sources to prognostic performance, we built six modality-specific systems on the same cohort ( n = 182) and compared them under 5-fold cross-validation (Figure 1b). The modalities included genomics (G), radiomics extracted from CT (P), DenseNet-derived histopathology features (D), and three fusion strategies—early fusion (EF), intermediate fusion (IF) and late fusion (LF). The validation results (Figure 2) reveal pronounced performance heterogeneity across folds. In folds 1-3, all single-modality models achieved only modest discrimination (C-index 0.65-0.72), and the fusion models did not rescue this deficit—likely because substantial distribution shifts between training and test splits were exacerbated by the limited sample size. By contrast, fold 4 exhibited marked improvements, with genomics (C-index = 0.81) and DenseNet features (0.83) outperforming other modalities, while fold 5 showed uniformly strong accuracy (C-index > 0.85) across all models. Importantly, the benefit of multimodal integration was fold-dependent: in fold 4, early fusion of G and D boosted the C-index by 10 % (0.83 → 0.91), whereas the same strategy yielded only a 2 % gain (0.88 → 0.90) in fold 5. This suggests that early fusion can exploit complementary signals when modalities convey consistent prognostic information, but may default to cautious, intermediate predictions when late-time survival signals conflict (e.g., ≈1000 days post-surgery in fold 5). No single feature type dominated across all folds (maximum inter-fold C-index variation ± 0.15), underscoring that robust risk stratification requires synergistic integration of complementary data modalities rather than reliance on any single source.
Figure 2: A comparative analysis of the C-index for linear and nonlinear models for six distinct modalities—genomics (G), pyradiomics (P), DenseNet (D), early fusion (EF), intermediate fusion (IF), and late fusion (LF)—is presented. Using CoxNet, features of varying dimensionality—50-dimensional for genomics (G), 107-dimensional for pyradiomics (P), and 1024-dimensional for DenseNet (D)—are directly input to establish baseline performance for each modality.
3.3 IDLE Score Contributions to TNM Staging and Tumor Grade
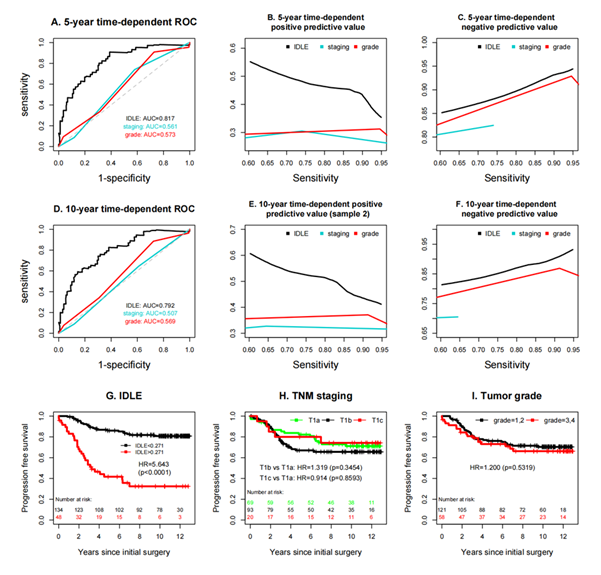
Because models that exploit complementary information outperform those built on a single feature type, we integrated the CT-derived radiomic features with the H&E-based histopathologic features to generate the final IDLE score (Figure 1b, c). The integration of IDLE scores significantly enhanced predictive accuracy, as illustrated in Figure 3 a–i. Specifically, the 5-year time-dependent ROC curve (Figure 3a) shows an AUC for IDLE of 0.817 ± 0.037, markedly higher than the AUCs for tumor grade (0.561 ± 0.042) and TNM staging (0.574 ± 0.044). Similarly, the 10-year time-dependent ROC curve (Figure 3d) indicates an AUC for IDLE of 0.792 ± 0.039, outperforming tumor grade (0.507 ± 0.041) and TNM staging (0.569 ± 0.045). Due to constraints in achieving higher sensitivities, the 5-year and 10-year time-dependent negative predictive values (NPVs) for TNM staging (Figures 3c, f) were evaluated only at sensitivities below 0.74 and 0.64, respectively. Across a sensitivity range of 60–95%, the 5-year and 10-year time-dependent positive predictive values (PPVs) for IDLE (Figures 3b, e) were consistently superior, while its NPVs (Figures 3c, f) were inferior compared to TNM staging and tumor grade. Additionally, survival analyses (Figure 3g–i) further highlight IDLE's prognostic value, with Figure 3g showing a hazard ratio (HR) of 5.643 (p<0.0001) for IDLE. Figure 3h demonstrating significant differences in progression-free survival across TNM stages (e.g., T1b vs. T1a: HR=1.319, p=0.3454), and Figure 3i indicating a higher risk for grade 3–4 tumors (HR=1.200, p=0.5519).
The single-dataset positive predictive values (PPVs, Figure 3b, e), though superior to TNM staging and tumor grade, exhibit greater variability across sensitivity ranges (60–95%) compared to the more stable PPVs from multiple datasets, which benefit from broader data representation. Additionally, the hazard ratio for IDLE in the single-dataset survival analysis (Figure 3g: HR=5.643, p<0.0001). However, no single input variable stood out and none of the individual features was sufficient to serve as a standalone marker. Even when combining multiple LDCT image features or various histopathological morphological features, the predictive accuracy of these methods was significantly lower than that of merging both LDCT and histopathology image features.
The findings further indicate that the IDLE score adds significant value beyond TNM staging and tumor grade, showing a strong association with cancer progression (Table 2), even when adjusted for these traditional factors. These results underscore the ability of IDLE to capture risk factors for cancer progression that are not reflected by TNM staging and tumor grade alone.
Figure 3: An evaluation of the predictive precision among IDLE, TNM classification, and tumor grade. Since higher sensitivities were unattainable, the negative predictive values for TNM classification could only be analyzed within a limited range of sensitivity levels with moderate values. The Kaplan–Meier curves in plot I did not include patients with an undetermined (GX) tumor grade.
3.4 IDLE Multivariate Analysis
Table 2: Multivariate analysis to study the added value of IDLE
|
HR |
95%CI |
||
|
IDLE high |
5.6708 |
(3.1650,10.1605) |
<0.0001 |
|
0.8665 |
(0.4667,1.6087) |
0.6499 |
|
|
0.7708 |
(0.2620,2.2680) |
0.6364 |
|
|
High grade |
0.9818 |
(0.5440,1.7720) |
0.9513 |
|
Age at surgery |
1.0318 |
(0.9738,1.0934) |
0.2888 |
|
Chemotherapy |
0.67 |
(0.2806,1.5996) |
0.3671 |
|
Radiotherapy |
1.3959 |
(0.4064,4.7945) |
0.5963 |
The reference is the T1a subgroup
4. Discussion
In this study, we demonstrated that integrating comprehensive tumor characteristics from preoperative low-dose CT scans with histological details from H&E-stained tissue images, processed through deep learning model, provides a more accurate prediction of the aggressiveness of stageT1A non-small cell lung cancers compared to traditional reliance on TNM staging and tumor grade alone. The IDLE method outperformed these conventional approaches, as evidenced by consistently higher AUC values, positive predictive values, and negative predictive values. These results suggest that the IDLE score holds promise as a tool for identifying high-risk patients who may experience cancer progression shortly after primary surgery and for selecting candidates who may benefit from early intervention.
By analyzing the global image characteristics from LDCT, the reasons behind the synergy between these global features and local tissue attributes within the deep learning model were uncovered. However, by integrating these hidden layer variables, the model could differentiate between patients with cancer progression and those without. Among the variables utilized by the deep learning network, no single input variable stood out; in other words, none of the individual features was sufficiently robust to serve as a standalone marker. Even when combining multiple LDCT image features or various histopathological morphological features, the predictive accuracy of these methods was significantly lower than that of IDLE, which merged both LDCT and histopathology image features.
Integrating LDCT and histology characteristics increase predictive accuracy, as standardized tissue histopathological characteristics allow insights of the local properties of tumors [40-42]. However, the histological analysis of the tumor tissue alone is insufficient to fully comprehend the tumor's interaction with the overall lung environment [34]. Due to the fact that LDCT image features quantify global tumor morphology [51], and H&E quantifies local tumor morphology [43], characteristics of the tumor from these two feature platforms usually do not overlap. This finding indicates that integrating characteristics from distinct, non-overlapping platforms yields a significantly more accurate predictor compared to merging multiple features from a single platform with potentially redundant data [44]. Additionally, the prediction accuracy may be further enhanced with integration of molecular biomarkers with IDLE.
Notably, there was an inverse relationship between progression-free survival and the duration from the final low-dose CT (LDCT) scan to the surgical procedure. This interval was identified as a critical factor by the deep learning model. A longer gap between LDCT screening and surgery indicated a delayed cancer diagnosis, which was observed in 21 out of the 54 patients who experienced disease progression (Table 1). Those cancers were not only still in stage IA, but were also of a higher grade. This can be explained by potential higher grades of the tumors (biologically more aggressive) and the early surgical intervention might have prevented or delayed progression. Timely diagnosis of aggressive lesions is certainly a requirement for improving the screening effectiveness of lung cancer.
In addition, multiple early and late fusion techniques [45-47], when applied with the Cox proportional hazards model, hold promise for predicting recurrence. Various modalities are shown to be better at different times, which furthermore suggests that predicting behavior is improved by combining multiple modalities. No single one was found to be better than the rest over all folds [47], as neural network methods are more difficult to generalize when trained on a limited quantity of data.
Multimodal fusion networks are a potentially fascinating area for future study; however, transfer learning from single modality datasets could enhance the training of these networks [49]. Multistage fusion simulating biological interactions between imaging and genomics is also of interest to investigations [50,51].
This research is constrained by its relatively limited sample size, with findings corroborated solely through cross-validation. Further validation from a more extensive cohort is necessary [45]. Although limitations exist, this investigation supports the initial hypothesis that integrating diverse tumor morphological characteristics from various imaging modalities [46] enhances the prediction of lung cancer progression risk, surpassing the capabilities of TNM staging and tumor grade alone.
5. Conclusion
To predict lung cancer recurrence, the study developed a deep learning model. Patient demographics, type of surgery, residual disease after surgery, lymph node dissection received, surgical tissue associated features, preoperative LDCT lung image features and interval between preoperative LDCT were analyzed to calculate the IDLE score. The second part of the study highlighted the potential of multimodal fusion, using both early and late fusion techniques with the Cox proportional hazards model, to predict recurrence. Different modalities work better in different contexts, indicating space to improve prediction by fusing information across multiple modalities. However, there is no single method which outperforms the other methods in all the folds, thus training neural networks on a small dataset remains difficult to generalize. This approach might facilitate the training of multimodal fusion networks through transfer learning from unimodal datasets.
Declaration
Funding
Funding is not available for this manuscript.
Data Availability
Data is provided within the manuscript or supplementary information files.
References
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 73 (2023): 17–48.
- Malhotra J, Malvezzi M, Negri E, et al. Risk factors for lung cancer worldwide. Eur Respir J 48 (2016): 889–902.
- Polanski J, Jankowska-Polanska B, Rosinczuk J, et al. Quality of life of patients with lung cancer. OncoTargets Ther 9 (2016): 1023–1028.
- Tang Y, Qiao G, Xu E, et al. Biomarkers for early diagnosis, prognosis, prediction and recurrence monitoring of non-small-cell lung cancer. OncoTargets Ther 10 (2017): 4527–4534.
- Fujinami R, Otis-Green S, Klein L, et al. Quality of life of family caregivers and challenges in caring for patients with lung cancer. Clin J Oncol Nurs 16 (2012): E210–E220.
- Pompili C. Quality of life after lung resection for lung cancer. J Thorac Dis 7 (2015): S138–S144.
- Fedor D, Johnson WR, Singhal S. Local recurrence after lung-cancer surgery: incidence, risk factors and outcomes. Surg Oncol 22 (013): 156–161.
- Taylor MD, Nagji AS, Bhamidipati CM, et al. Tumour recurrence after complete resection for non-small-cell lung cancer. Ann Thorac Surg 93 (2012): 1813–1821.
- Tachibana T, Matsuura Y, Ninomiya H, et al. Optimal treatment strategy for oligo-recurrence lung-cancer patients with driver mutations. Cancers 16 (2024): 464.
- Li L, Zhou T, Xu Y et al. Recurrence/prognosis estimation using a molecularly positive surgical-margin model in pIIIA/N2 NSCLC. Mol Oncol 18 (2024): 1649-1664.
- Grambozov B, Wass R, Stana M, et al. Impact of re-irradiation, chemotherapy and immunotherapy on survival in recurrent lung cancer. Thorac Cancer 12 (2021): 1162–1170.
- Bucknell NW, Hardcastle N, Bressel M, et al. Pulmonary function in a randomised trial of single- versus multi-fraction SBRT for pulmonary oligometastatic disease (TROG 13.01 SAFRON II). Int J Radiat Oncol. Biol Phys 114 (2023): 944-951.
- Li Y, Liu L, You R, et al. Effect of initial recurrence site on prognosis of non-small-cell lung-cancer subtypes: a retrospective cohort study. World J Surg Oncol 21 (2023): 360.
- Li Y-Z, Kong S-N, Liu Y-P, et al. Can ctDNA/cfDNA liquid biopsy replace tissue biopsy for precision treatment of EGFR-mutated NSCLC? J Clin Med 12 (2023): 1438.
- Pessoa LS, Heringer M, Ferrer VP. ctDNA as a cancer biomarker: a broad overview. Crit Rev Oncol Hematol 155 (2020): 103109.
- Balloux F, Brynildsrud OB, Dorp LV, et al. From theory to practice: translating whole-genome sequencing into the clinic. Trends Microbiol 26 (2018): 1035–1048.
- Cuppen E, Elemento O, Richard Rosenquist R, et al. Implementation of whole-genome and transcriptome sequencing into clinical cancer care. JCO Precis Oncol 6 (2022): e2200245.
- Huang P, Lin CT, Li Y, et al. Prediction of lung-cancer risk at follow-up screening with low-dose CT by deep learning. Lancet Digit. Health 1 (2019): e353–e362.
- Bortolotto C, Pinto A, Brero F, et al. CT and MRI radiomic features of lung cancer: comparison and software consistency. Eur Radiol Exp 8 (2024): 1.
- Chen J-M, Li Y, Xu J, et al. Computer-aided prognosis on breast cancer with H&E images: a review. Tumour Biol 39 (2017): 1010428317694550.
- Lee S, Jung J, Hong H, et al. Radiomic feature-based prediction model of lung-cancer recurrence in NSCLC patients. Proc SPIE 11515 (2020): 115150N.
- Aonpong P, Iwamoto Y, Wang W, et al. Hand-crafted and deep-learning radiomics models for NSCLC recurrence prediction. In Innovation in Medicine and Healthcare (2020): 141–151.
- Webb S. Deep learning for biology. Nature (2018).
- Wang X, Janowczyk A, Zhou Y, et al. Prediction of recurrence in early-stage non-small-cell lung cancer using nuclear features from digital H&E images. Sci Rep 7 (2017): 13543.
- Mohamed SK, Walsh B, Timilsina M, et al. On predicting recurrence in early-stage non-small-cell lung cancer. AMIA Annu Symp Proc 2021 (2022): 853–862.
- Jimenez C, Ma J, Gonzalez AR, et al. TNM staging and overall survival in patients with pheochromocytoma and sympathetic paraganglioma. J Clin Endocrinol Metab 108 (2023): 1132–1142.
- Oshikiri T, Hironobu G, Takashi K, et al. Proposed modification of the eighth edition of the AJCC-ypTNM staging system of esophageal squamous cell cancer treated with neoadjuvant chemotherapy: unification of the AJCC staging system and the Japanese classification. Eur J Surg Oncol 48 (2022): 1760–1767.
- Guillen G. Digital Image Processing with Python and OpenCV (2019): 97–140.
- Dejima H, Iinuma H, Kanaoka R, et al. Exosomal microRNA in plasma as a biomarker for NSCLC recurrence. Oncol Lett 13 (2017): 1256–1263.
- Patz EFJr, Caporaso NE, Dubinett SM, et al. NLST/ACRIN 6654 biorepository: design and specimen availability. J Thorac Oncol 5 (2010): 1502–1506.
- Huang P, Park S, Yan R, et al. Added value of computer-aided CT features for early lung-cancer diagnosis with small nodules. Radiology 286 (2018): 286–295.
- Aerts HJWL, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype via quantitative radiomics. Nat Commun 5 (2014): 4006.
- Passiglia F, Bronte G, Castiglia M, et al. Prognostic and predictive biomarkers for targeted therapy in NSCLC. Expert Opin Biol Ther 15 (2015): 1553–1566.
- van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 77 (2017): e104–e107.
- Bisong E. The multilayer perceptron (MLP). in Building Machine Learning and Deep Learning Models on Google Cloud Platform (2019): 401–405.
- Cox DR. Regression models and life-tables. J R Stat Soc B 34 (1972): 187–202.
- Durisová M, Dedík L. SURVIVAL—software for survival-curve estimation and comparison. Methods Find Exp Clin Pharmacol 15 (1993): 535–540.
- Pölsterl S. scikit-survival: a library for time-to-event analysis built on top of scikit-learn. J Mach Learn Res 21 (2020): 1–6.
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2021).
- Friedman J, Hastie T, Tibshirani R. Regularization paths for GLMs via coordinate descent. J Stat Softw 33 (2010): 1–22.
- Detterbeck FC, Boffa DJ, Kim AW, et al. The eighth-edition lung-cancer stage classification. Chest 151 (2017): 193–203.
- Camidge DR, Doebele RC, Kerr KM. Predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 16 (2019): 341–355.
- Mandrekar JN. Receiver-operating-characteristic curves in diagnostic tests. J Thorac Oncol 5 (2010): 1315–1316.
- Bakr S, Gevaert O, Echegaray S, et al. A radiogenomic dataset of non-small-cell lung cancer. Sci Data 5 (2018): 180202.
- Huang P, Illei PB, Franklin W, et al. Lung-cancer recurrence-risk prediction through integrated deep-learning evaluation. Cancers 14 (2022): 4150.
- Kroschke, J, von Stackelberg O, Heussel CP, et al. Imaging biomarkers in thoracic oncology: advances in radiomics for lung-cancer therapy response. RöFo (2022).
- Pölsterl S, Navab N, Katouzian A. Fast training of support-vector machines for survival analysis. In Machine Learning and Knowledge Discovery in Databases 9285 (2015): 243–259.
- Pronzato L, Rendas M-J. Weighted leave-one-out cross-validation. SIAM/ASA J. Uncertain. Quantif 12 (2024): 1213–1239.
- National Lung Screening Trial Research Team et al. Reduced lung-cancer mortality with low-dose CT screening. N Engl J Med 365 (2011): 395–409.
- Pairolero PC, Williams DE, Bergstralh EJ, et al. Postsurgical stage I bronchogenic carcinoma: morbid implications of recurrent disease. J Thorac Cardiovasc Surg 88 (1984): 911–918.
- Kanaoka R, Iinuma H, Dejima H, et al. Plasma exosomal microRNA-451a as a biomarker for early recurrence and prognosis of NSCLC. Oncology 94 (2018): 311–323.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 77.66%
Acceptance Rate: 77.66%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 