DNA Contamination During Extraction of Dried Blood Spots from Whatman Filter Paper
Hamma MAIGA*,1,2, Patrick J. GORRES1 and Patrick DUFFY1
1Laboratory of Malaria Immunology and Vaccinology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 29 Lincoln Drive, Bethesda, MD 20892, USA.
2Institut National de Santé Publique, Bamako, 1771, Mali
*Corresponding author: Hamma MAIGA, Laboratory of Malaria Immunology and Vaccinology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 29 Lincoln Drive, Bethesda, MD 20892, USA.
Received: 25 June 2025; Accepted: 05 July 2025; Published: 09 September 2025
Article Information
Citation: Hamma MAIGA, Patrick J. GORRES and Patrick DUFFY. DNA Contamination During Extraction of Dried Blood Spots from Whatman Filter Paper. International Journal of Applied Biology and Pharmaceutical Technology. 16 (2025): 30-34.
View / Download Pdf Share at FacebookAbstract
Dried Blood Spot (DBS) testing on confetti is one of the most common methods used to collect and extract DNA from field samples for molecular epidemiology of malaria studies. Hence, the present study aimed to investigate whether ethanol or other solutions used to clean scissors are a source of sample contamination. DBS were prepared from 3 drops of blood from two Plasmodium falciparum cultures spotted on confetti, and 3 solutions (water, ethanol, and DNase) were used to clean scissors between spots during DNA extraction. For each cleaning solution, two blank confetti were used as negative controls. Samples were analyzed by PCR-based genotyping of merozoite surface proteins 1 & 2 (msp-1 and msp-2), and P. falciparum chloroquine resistance transporter (Pfcrt). A total of 15 samples were analyzed. Based on msp-1 and msp-2 amplification, P. falciparum was detected on blank confetti when scissors were washed with ethanol. Based on Pfcrt amplification, DNA of P. falciparum was detected on blank confetti when scissors were cleaned with ethanol or water. No P. falciparum genes were detected on blank confetti when scissors were cleaned with DNase. Scissors cleaned with DNase prevented cross-contamination between samples during processing of DBS, whereas ethanol (which is commonly used) fails to avert cross-contamination.
Keywords
<p>DNA, Extraction, Scissors, DBS, DNase</p>
Article Details
Introduction
Malaria is a serious global health problem causing over 600,000 deaths in 2021 [9]. Correct diagnosis is necessary for prevention, control, and suitable treatment of malaria, commonly performed by microscopy, rapid diagnostic tests (RDTs), and molecular methods [6]. Success of a molecular method for malaria diagnosis depends critically on the quality of genomic DNA being used for examination, which in turn depends on accuracy of the employed isolation procedure. Multitudes of different DNA extraction methods have been published [7] since the first DNA isolation in 1869 [2], and each method must overcome specific extraction problems such as DNA shearing, background contamination, low purity, and low yield. Of these, Dried Blood Spot (DBS) is one of the most common methods used to collect and extract DNA in field studies of malaria molecular epidemiology, where transport and storage conditions are often suboptimal. This technique is minimally invasive (requiring only microliters of typically peripheral blood), inexpensive, and easy to multiplex and automate [5]. DBS samples are compatible with many bioanalytical methods, among them chromatography, mass spectrometry, DNA, and immunoassays. However, DNA extraction is particularly challenging in resource-limited regions, where scissors are used to cut filter papers, possibly introducing contamination between samples due to poor scissor cleaning.
Many DBS methods use 70% ethanol to clean the scissors, but it is unknown whether ethanol or other cleaning solutions may be a source of sample contamination. DNase is an endonuclease that digests single and double-stranded DNA; it hydrolyzes phosphodiester bonds producing mono- and oligodeoxyribonucleotides with 5’-phosphate and 3’OH groups. The enzyme activity is strictly dependent on Ca2+ and is activated by Mg2+ or Mn2+ ions: 1) in the presence of Mg2+, DNase cleaves each strand of dsDNA independently in a statistically random fashion [8]; 2) In the presence of Mn 2+, the enzyme cleaves both. DNA strands at approximately the same site, producing DNA fragments with blunt ends or with one or two nucleotide overhangs [8] It is in that light the present study aimed to investigate the contamination of samples extracted by scissors which may impact interpretation of molecular epidemiology results.
Material And Methods
DNA extraction from dried blood spot (DBS)
DBS were prepared from 3 drops of blood from the same culture, spotted on Whatman FTA cards (Whatman Inc., Brentford, UK), then dried and stored for 24 hours at room temperature (Figure 1). DBS on cards were cut into small pieces with scissors and transferred into 1.5 mL microtubes. Parasite DNA was extracted using QIAamp DNA blood mini kits (Qiagen, Valencia, CA USA) according to the manufacturer’s instructions and final eluted volume was 30 mL. DNA samples were stored at −20°C until use. The concentration of DNA was achieved using a Nano Drop 2000. DBS were prepared from 3 drops of blood from two Plasmodium falciparum cultures (20-0119-0 and 20-2091-0 isolates) spotted on filter paper (Figure 2). All samples were dried for 24 hours at room temperature. Three solutions (water, ethanol, and DNase) to clean scissors between spots during DNA extraction were examined. For each cleaning solution, two blank confetti were used as negative controls. Residual water (RW), ethanol (RE) and DNase (RD) after scissor washing were also amplified.
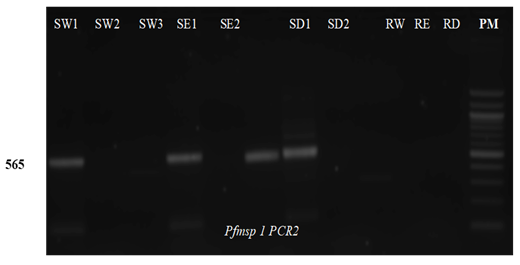
SW1 (a) is the reference with blood and SW2 and SW3 (b) are blank confetti catted after washing scissors with water the first and second time, respectively. (c-d) SE1 is the reference with blood and SE2 and SE3 are blank confetti catted after washing scissors with ethanol the first and second time, respectively. (e-f) SD1 is the reference with blood and SD2 and SD3 are blank confetti catted after washing scissor with DNase the first and second time, respectively.
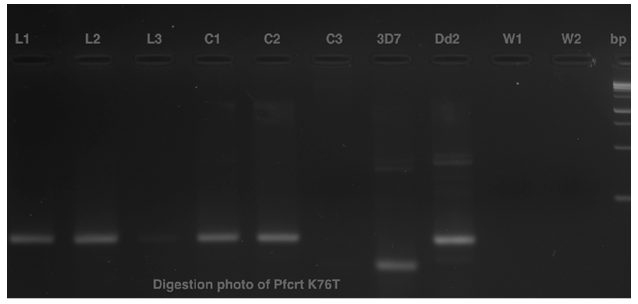
L1 and L2 are the reference with blood and C1 and C2 are blank confetti catted after washing scissors with ethanol the first time, respectively; L3 is the reference with blood and C3 is blank confetti catted after washing scissors with DNase the first time.
PCR assay of dried blood spot (DBS)
Parasite-infected and negative control samples were analyzed by PCR-based genotyping of P. falciparum merozoite surface proteins 1 and 2 (msp-1 and msp-2), and P. falciparum chloroquine resistance transporter (Pfcrt) genes. A total of 15 samples were analyzed. Based on msp1 and msp2 amplification (3 for each cleaning solution, n=9), P. falciparum was detected on blank confetti when scissors were washed with ethanol. Based on Pfcrt amplification (2 for each cleaning solution, n=6), DNA of P. falciparum was detected on blank confetti when scissors were washed with ethanol or water. Genotyping using amplification of msp-1 and msp-2 markers were performed on paired samples obtained. PCR primers sequences and cycles have been previously described [4]. Gels were stained with ethidium bromide and visualized under UV illumination. Band sizes of PCR products across the two markers were measured visually and compared. PCR products visualized on 1.5% agarose gel in 1×Tris-acetate-EDTA (TAE) and stained with ethidium bromide (Sigma-Aldrich, Sydney, NSW, Australia). Length differences were determined using a 100 bp molecular weight ladder (Promega). Electrophoresis conditions were 100 mV for 30 min. Allele-specific positive controls and DNA-free negative controls were included in each round of reactions. Primers for msp-1 and msp-2 were synthesized by Integrated DNA Technologies, Coralville, IA, USA.
Results
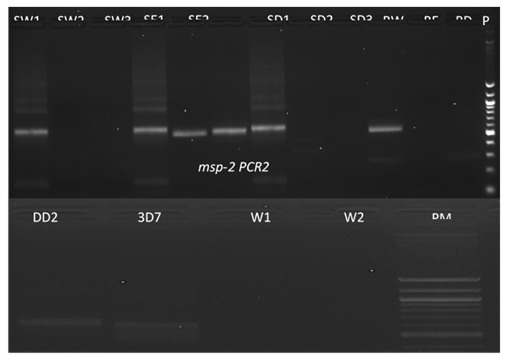
Data of the tested PCR revealed that no bands were detected in samples with scissors washed with DNase. Schematic representation of agarose gel electrophoresis of nested PCR products from SW1, SE1 and SD1 show the presence of bands at 565 bp and 534 bp in msp1 (Figure 3) and msp-2 (Figure 4) respectively. In the nested PCR assay using msp-1 and msp-2, no band was detected in SW1, SW2, SE2, SD1, SD2, RE and RD tested. Bands were detected in SE2 in msp-1 and SE1 and SE2 in msp-2, respectively. Only blank confetti catted by scissors washed with ethanol presented bands, detected in C1 and C2 on agarose gel electrophoresis of digestion PCR products, but no band was detected with C3 (Figure 5).
DBS catted by scissors washed with water (SW1 as reference, SW2 and SW3 from blank confetti); DBS catted by scissors washed with ethanol (SE1, SE2 and SE3) and DBS catted by scissors washed with DNase (SD1, SD2 and SD3). RW, RE and RD represent residual water, ethanol, and DNase, respectively.
DBS catted by scissors washed with water (SW1 as reference, SW2 and SW3 from blank confetti); DBS catted by scissors washed with ethanol (SE1 as reference, SE2 and SE3) and DBS catted by scissors washed with DNase (SD1 as reference, SD2 and SD3). RW, RE and RD represent residual water, ethanol, and DNase, respectively. DD2 and 3D7 are positive controls and water (W1 and W2) is negative control; PM is molecular marker.
Pfcrt products from the digestion run on a 1.5% agarose gel. 100 bp molecular weight ladder (Sigma P1473). DBS catted by scissors washed with ethanol (L1, L2 as reference, C1 and C2) and DBS catted by scissors washed with DNase (L3 as reference, C3). W1, W2, 3D7 and Dd2 represent controls, respectively.
Discussion
Dried blood spots (DBS) typically consist in the deposition of small volumes of capillary blood onto dedicated paper cards. DBS samples are simple to prepare and transport, enabling safe and accessible diagnostics, both locally and globally. DBS can provide sufficient sample stability of at least a week for many relevant analytes, allowing ambient temperature transportation to a centralized specialist laboratory anywhere in the world. Today, thousand and thousands of filter paper are processed in many malaria studies with particular importance for public health perspective. Thus, we think this report will add important information’s in the field. We have assessed water, ethanol and DNAse to clean scissors during the filter paper cutting process. Ethanol is a non-additive precipitant fixative that shifts isoelectric points of proteins less than other fixatives [3]. It fixes proteins by dehydration and precipitation, the degree of which being dependent on the amount of water present and the solubility of materials in the mixture. The basic aims of fixation are to preserve the tissue nearest to its living state, and to prevent any change in shape and size of tissue at the time of processing. This could possibly explain how pieces of blood-stained confetti attached to scissors after being dipped in ethanol. In addition, we observed one band from residual of water (RW) in msp-2 for representing when observed. Water is by far the most studied chemical compound [1] and called the universal solvent since it can dissolve more substances than any other liquid. After dipping in water, particles of confetti on scissors detached themselves and remained at the bottom of the water. We did not detect any band with msp-1 and msp-2 in scissors washed with DNase. Although these methods may suit to LMICs with limited resources, more elaborate methods exist now and prevent any contact with the DBS; it uses laser to cut DBS avoiding any hand contact and hence contamination. But these methods are expensive for low-income countries.
Conclusion
Scissors cleaned with DNase prevented cross-contamination between samples during processing of dried blood spots. Ethanol cleaning of scissors, which is commonly used, failed to avert cross-contamination. Sample cross-contamination can reduce diagnosis accuracy; we recommend DNase wash as the standard cleaning solution to improve DNA extraction.
Competing Interests
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank Justin Yai Doritchamou of LMIV for providing the blood sample, and the NIH Fellows Editorial Board for critical editing.
Authors Contribution
HM participated in the study design, collection, analysis and wrote the first draft of the manuscript. PJG and PD edited the manuscript.
Funding
This work supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. Hamma Maiga was the recipient of African Postdoctoral Training Initiative (APTI) Scholarship (#APTI-18-01#). The content is solely the responsibility of the authors.
References
- Akiya N, Savage PE. 2002. Roles of Water for Chemical Reactions in High-Temperature Water. Chemical Reviews, 102(8), 2725–2750. DOI: https://doi.org/10.1021/cr000668w
- Dahm R. 2008. Discovering DNA: Friedrich Miescher and the early years of nucleic acid research. Human Genetics, 122(6), 565–581. DOI: https://doi.org/10.1007/s00439-007-0433-0
- De PR Y. 2006. Biological Techniques. Discovery publishing House.
- De Radiguès X, Diallo KI, Diallo M, Ngwakum PA, Maiga H, Djimdé A, Sacko M, Doumbo O, Guthmann, JP. 2006. Efficacy of chloroquine and sulfadoxine/pyrimethamine for the treatment of uncomplicated falciparum malaria in Koumantou, Mali. Transactions of the Royal Society of Tropical Medicine and Hygiene, 100(11), 1013–1018. DOI: https://doi.org/10.1016/j.trstmh.2006.03.004
- Demirev PA. 2013. Dried Blood Spots: Analysis and Applications. Analytical Chemistry, 85(2), 779–789. DOI: https://doi.org/10.1021/ac303205m
- Fontecha GA, Mendoza M, Banegas E, Poorak M, De Oliveira AM, Mancero T, Udhayakumar V, Lucchi NW, Mejia RE. 2012. Comparison of molecular tests for the diagnosis of malaria in Honduras. Malaria Journal, 11(1), 119. DOI: https://doi.org/10.1186/1475-2875-11-119
- Kuhn R, Böllmann J, Krahl K, Bryant IM, Martienssen M. 2017. Comparison of ten different DNA extraction procedures with respect to their suitability for environmental samples. Journal of Microbiological Methods, 143, 78–86. DOI: https://doi.org/10.1016/j.mimet.2017.10.007
- Sambrook J, Russell D. 2001. Molecular Cloning: A Laboratory Manual (the third edition). Cold Spring Harbor Laboratory Press.
- 2023. World Malaria Report. WHO, editor. Geneva, Switzerland: World Health Organization.




 Impact Factor: * 3.0
Impact Factor: * 3.0 Acceptance Rate: 76.32%
Acceptance Rate: 76.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
