Novel Candida sequence types and antifungal profiles in a Northern Brazil Hematology Center
Cristina Motta Ferreira*,1,2, Guilherme Motta Antunes Ferreira3, Larissa Alves Gomes4, Adryele Ramos Pazini4, Victor Costa de Souza3, Joseir Saturnino Cristino1, George Allan Villarouco da Silva2, Andrea Monteiro Tarragô2, 1Franceline Oliveira Calheiros, Felipe Gomes Naveca3, William Antunes Ferreira2
1Hospital Foundation of Hematology and Hemotherapy of Amazon- HEMOAM.
2Pos-Graduate Program of Applied Sciences in Hematology. Amazon, Brazil.
3Leônidas and Maria Deane Institute - ILMD/FIOCRUZ, Graduate Program in Biology of Pathogen-Host Interaction. Amazon, Brazil.
4FAPEAM-Research Support Foundation of the State of Amazon. Amazon, Brazil.
*Corresponding author: Cristina Motta Ferreira, Amazonas Hematology and Hemotherapy Hospital Foundation - HEMOAM, Av. Constantino Nery, 4397, Chapada, Postal Code: 69050-001. Manaus, Amazonas, Brazil.
Received: 04 November 2025; Accepted: 11 November 2025; Published: 24 November 2025
Article Information
Citation: Cristina Motta Ferreira, Guilherme Motta Antunes Ferreira, Larissa Alves Gomes, Adryele Ramos Pazini, Victor Costa de Souza, Joseir Saturnino Cristino, George Allan Villarouco da Silva, Andrea Monteiro Tarragô, Franceline Oliveira Calheiros, Felipe Gomes Naveca, William Antunes Ferreira. Novel Candida sequence types and antifungal profiles in a Northern Brazil Hematology Center. Archives of Microbiology and Immunology. 9 (2025): 252-259.
View / Download Pdf Share at FacebookAbstract
Introduction: Invasive fungal infections, particularly candidiasis, remain a major cause of morbidity and mortality in patients with hematologic malignancies, complicated by emerging antifungal resistance.
Methods: Here we characterized Candida species isolated from hematologic patients at a tertiary referral center in Northern Brazil using antifungal susceptibility testing and multilocus sequence typing.
Results: We identified C. albicans, C. krusei, and C. tropicalis isolates, including fluconazole and amphotericin B resistant C. krusei strains. Molecular analysis revealed three novel C. albicans sequence types and a new C. krusei sequence type, phylogenetically linked to global circulating clones. These findings indicate local revolution and potential clonal dissemination within the hospital environment.
Conclusion: Our study underscores the importance of ongoing molecular surveillance and antifungal susceptibility monitoring to inform infection control and optimize treatment strategies in immunocompromised hematologic patients. This is the first report of novel Candida sequence types in a hematologic reference center in the Amazon region.
Keywords
Molecular epidemiology, invasive fungal infection, oncohematological patient, Candida infections, Hematologic malignancies, antifungal resistance
Molecular epidemiology articles, invasive fungal infection articles, oncohematological patient articles, Candida infections articles, Hematologic malignancies articles, antifungal resistance articles
Article Details
Introduction
Candida albicans is a commensal yeast that is part of the normal human microflora. This species can also act as an infectious agent, causing invasive disease, especially in oncohematological patients such as those with acute lymphocytic leukemia (ALL), acute myeloid leukemia (AML), or those undergoing chemotherapy [1-5]. Invasive candidiasis remains a significant concern in hospitals worldwide, with its incidence increasing in recent years [5,6]. The epidemiological profile of candidemia varies among cancer patients, particularly those with oncohematologic malignancies [5]. Invasive fungal infection (IFI) poses a significant threat to oncohematological patients and is one of the main causes of morbidity and mortality [1,7]. Clinical signs are often nonspecific because of the lack of characteristic symptoms. Furthermore, resistance to standard antifungal treatments leads to longer hospital stays, treatment difficulties, and higher healthcare costs [1,7]. Among the yeasts, Candida albicans (C. albicans) and non-Candida albicans (NAC) species – including Candida glabrata (C. glabrata), Candida tropicalis (C. tropicalis), Candida parapsilosis (C. parapsilosis), and Candida krusei (C. krusei) – account for more than 90% of invasive candidiasis cases [4]. The reported incidence ranges from 0,15% to 1,5% in hospitalized patients with underlying malignancies [6]. In recent years, emerging infections caused by NAC species have become an increasing clinical concern [8].
The incidence and mortality rates of IFIs vary by region. In Iran, the estimated mortality from Candida infections ranges from 30% to 80%, while in Taiwan it is approximately 5.9%. In India, the prevalence among AML and ALL patients was 62.5% (15/24) and 37.5% (9/24), respectively [9]. Vargas-Espindola et al. [6] reported a mortality rate of 50% in a multicenter study conducted at five cancer treatment centers in Bogota, which is similar to the global average found in various studies [10-12]. In Brazil, Camplesi Jr. et al. [1] identified invasive fungal infections in 16.2% (n = 19) of hematologic patients, with 6 cases caused by yeasts and 13 by filamentous fungi. In Manaus (Amazonas, Brazil), mycological diagnostic records from the Bacteriology Laboratory of the HEMOAM Hospital Foundation (January–August 2022) showed that fungal species isolated from patients with hematologic diseases included: C. albicans (2.5%), C. guilliermondii (21.0%), C. tropicalis (5.3%), C. pelliculosa (5.3%), C. famata (5.3%), C. krusei (8.0%), C. haemulonii (7.7%), C. parapsilosis (26.4%),Cryptococcus laurentii(13.2%), andFusariumspp. (5.3%). Even with various therapeutic options available, the prevalence of fungal infections has increased, possibly due to the use of antifungal drugs as prophylaxis, which may encourage the development of resistance mechanisms in these microorganisms. Additionally, current medications, such as polyenes (amphotericin B), azoles (itraconazole, fluconazole, voriconazole), and echinocandins (caspofungin, micafungin, and anidulafungin), still have drawbacks related to effectiveness, selectivity, toxicity, resistance, and spectrum of activity [13-15].
Therefore, due to the ongoing global reports of therapeutic failures in fungal infections caused by resistant strains and their potential link to higher mortality rates, the World Health Organization (WHO) published a list of priority fungal pathogens, including C. albicans in the critical group; C. tropicalis and C. parapsilosis in the high priority group [16]. There is an urgent need to implement susceptibility testing methods for fungi in hospitals, both to gather data for a better understanding of the issue and to identify the most effective treatments for patients [4]. In addition to resistance profiles, molecular epidemiology is crucial for understanding the evolutionary dynamics ofCandidaspecies and mapping the geographic distribution of resistant and non-resistant clonal groups. Several studies have identified dominant sequence types (STs), such as ST69 and ST90 forC. albicansand ST256 forC. krusei, which have been isolated from various human and animal biological samples across continents, including Australia, Asia, Europe, North America, and South America (including Brazil) [2, 17, 18, 27]. Therefore, this study aims to investigate the infectious processes caused by differentCandidaspecies in hematological patients admitted to a reference hematology and hemotherapy hospital in northern Brazil. The study will evaluate the antifungal susceptibility profiles and molecular epidemiology of the isolates to determine their origin and evolutionary relationships among circulating sequence types. The findings may help develop strategies to prevent outbreaks, limit the spread of resistance, and improve clinical outcomes and prognosis for patients affected by this condition.
Methods
Study design and Patients
This is a qualitative, observational, and exploratory study with experimental elements, based on the analysis of five cases of infections caused byCandidaspp. The study used data from diagnostic and therapeutic procedures, electronic medical records, and laboratory records from the HEMOAM Hospital Foundation. The cases involved hematological patients of both genders and all age groups who were treated or hospitalized with suspected or confirmed fungal infections between January 2022 and December 2023. The cases described refer to patients diagnosed and treated at FHEMOAM, originating from the capital city of Manaus and rural municipalities in the State of Amazonas, including Eirunepé, Fonte Boa, São Gabriel da Cachoeira, Manacapuru, São Sebastião de Uatumã, and Santa Isabel do Rio Negro. These patients had previously been diagnosed with hematological cancers, including B-lineage acute lymphoblastic leukemia (ALL-B), acute myeloid leukemia (AML), diffuse large B-cell lymphoma, and high-risk Burkitt lymphoma. All patients were hospitalized for chemotherapy and developed neutropenia during treatment, along with clinical suspicion of infection. Blood and urine cultures were ordered, and microbiological tests identifiedC. albicans,C. tropicalis, andC. krusei. After identifying the fungal pathogens, antifungal therapy with fluconazole was started, leading to favorable outcomes in patients with large cell diffuse lymphoma, AML, and Philadelphia chromosome-positive ALL-B. The patients diagnosed with ALL-B and high-risk Burkitt lymphoma, however, died.
Microbiological Diagnosis
The samples were collected during routine procedures at the FHEMOAM Clinical Microbiology Laboratory. The biological samples sent to the bacteriology laboratory included urine and blood. Urine was cultured on URILAB-CLED/MacConkey slide culture media (Laborclin- Biokar Diagnostics, Allonne, France) for primary seeding. After 24 hours of incubation and a colony count of 1,000,000 CFU/mL, a subculture was performed on 5% sheep blood agar, Mueller-Hinton agar, MacConkey agar, and Sabouraud agar (Himedia, Hexasystems, Mumbai, India), followed by another 24 hours of incubation at 35.4°C. A pure colony grown on Sabouraud agar was selected for subsequent tests and Gram staining. The positive blood sample was cultured on 5% sheep blood agar, chocolate agar, Mueller-Hinton agar, MacConkey agar, and Sabouraud agar at 35.4°C for 24 hours. After incubation, a pure colony from Sabouraud agar was chosen for further testing and Gram staining. Using standard microbiological procedures, phenotypic identification and susceptibility testing (including antifungal susceptibility) with respective MIC values were performed using automated equipment (VITEK-2, bioMérieux, Craponne, France), following the manufacturer’s instructions and the 2024 guidelines from the Brazilian Committee on Antimicrobial Susceptibility Testing (BrCAST) and the Clinical & Laboratory Standards Institute (CLSI). Isolates ofC. albicans,C. tropicalis, andC. kruseidemonstrating resistance in antifungal susceptibility testing (antifungigram) were preserved at –80°C in cystine tryptic soy broth supplemented with 20% glycerol at the Bacteriology Laboratory of the HEMOAM Foundation.
Molecular Epidemiology
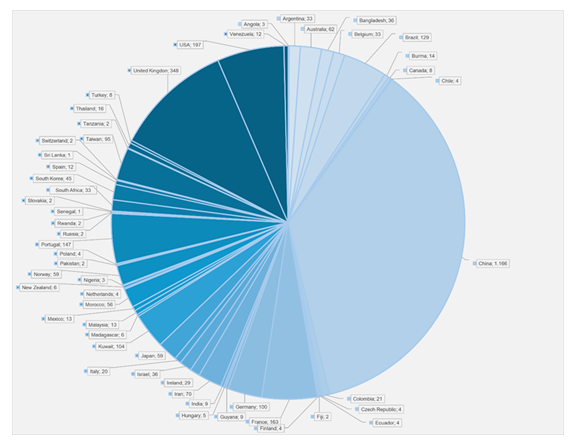
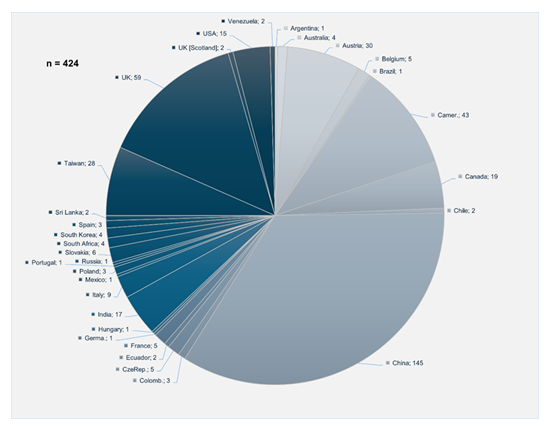
Genomic DNA fromCandidaspp. isolates was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen, CA, USA) following the manufacturer’s protocol, with minor modifications. Polymerase chain reaction (PCR) was performed to amplify multilocus sequence typing (MLST) genes specific to C. albicans,C. tropicalis, andC. krusei, following protocols described in previous studies [19-21]. PCR products were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) and analyzed with the ABI PRISM 3130 XL Genetic Analyzer (Applied Biosystems, Foster City, CA), according to the manufacturer’s instructions. Sequence analysis of each MLST gene forC. albicans,C. tropicalis, andC. kruseiwas conducted following protocols available on the PubMLST website (https://pubmlst.org/organisms) [22]. Sequence fragments were mapped, trimmed, and aligned using Geneious v.2025 software with reference sequences from the PubMLST database. Sequence typing was performed by comparing allele combinations with database entries to determine the sequence type (ST). Newly identified STs forC. albicansandC. kruseiwere uploaded to the MLST database. Phylogenetic models of the newly identified STs ofC. albicanswere constructed using allele profile data (AAT1a, ACC1, ADP1, SYA1, VPS13, and ZWF1b) from 3,252 entries, and forC. kruseiusing 427 entries (HIS3, LEU2, NMT1, TRP1, LYS2D), obtained from various sources (Table 1) and regions (Graphic 1 and Graphic 2). Notably, the only record forC. kruseiin the PubMLST database corresponds to the strain described in this study (Graphic 2). Phylogenetic trees were generated using Phyloviz v2.2software, available athttps://www.phyloviz.net/goeburst/ and https://www. phyloviz.net/ [23,24].
Table 1: Quantification of human biological samples used on the eBURST organogram
|
Source |
C. albicans |
C. krusei |
||
|
n |
% |
n |
% |
|
|
Blood |
1.295 |
39,8 |
154 |
56,4 |
|
Catheter |
47 |
1,4 |
- |
- |
|
Feces |
135 |
4,2 |
- |
- |
|
Bronchial lavage |
25 |
0,8 |
- |
- |
|
Other sterile sites |
- |
- |
28 |
10,3 |
|
Other superficial source |
- |
- |
43 |
15,8 |
|
Oropharynx |
579 |
17,8 |
- |
- |
|
Oropharynx/sputum |
- |
- |
39 |
14,3 |
|
Skin |
65 |
2,0 |
- |
- |
|
Sputum |
148 |
4,6 |
- |
- |
|
Urine |
209 |
6,4 |
- |
- |
|
SWAB vaginal |
726 |
22,3 |
11 |
4 |
|
Wound |
23 |
0,7 |
- |
- |
|
Not informed |
- |
- |
152 |
55,7 |
Results
Microbiological analyses identified three (3) isolates ofC. albicans, one (1)C. tropicalis, and two (2)C. krusei. The results of antifungal susceptibility testing (AST), including minimum inhibitory concentrations (MICs) and intermediate susceptibility phenotypes (i.e., susceptible, increased exposure), are shown in Table 3. Among the isolates,C. krusei showed resistance to fluconazole (MIC = 8 µg/mL) and amphotericin B (MIC=2 µg/mL).C. albicansexhibited an intermediate phenotype to voriconazole (MIC ≤ 0.12 µg/mL), whileC. tropicalis was susceptible to all tested antifungal agents. Susceptibility profiles ofC. albicansto the echinocandins caspofungin and micafungin could not be classified because clinical breakpoint values for these agents have not yet been established by the European Committee on Antimicrobial Susceptibility Testing (EUCAST v10.0) or the Clinical and Laboratory Standards Institute (CLSI, 2024) (Table 2).
Table 2: Candida spp. antimicrobial susceptibility test
|
Isolates |
id |
Antifungical |
MIC |
Interpretation |
|
Candida albicans |
10 |
Fluconazole |
≤0.5 |
S |
|
Voriconazole |
≤0.12 |
I |
||
|
Caspofungin |
≤0.12 |
- |
||
|
Micafungin |
≤0.06 |
- |
||
|
Amphotericin B |
1 |
S |
||
|
68 |
Fluconazole |
1 |
S |
|
|
Voriconazole |
≤0.12 |
I |
||
|
Caspofungin |
≤0.12 |
- |
||
|
Micafungin |
≤0.06 |
- |
||
|
Amphotericin B |
1 |
S |
||
|
126 |
Fluconazole |
2 |
S |
|
|
Voriconazole |
≤0.12 |
I |
||
|
Caspofungin |
≤0.12 |
- |
||
|
Micafungin |
≤0.06 |
- |
||
|
Amphotericin B |
0,5 |
S |
||
|
Candida krusei |
58 |
Fluconazole |
8 |
R |
|
Voriconazole |
≤0.12 |
IE |
||
|
Caspofungin |
0.25 |
- |
||
|
Micafungin |
0.12 |
IE |
||
|
Amphotericin B |
2 |
R |
||
|
Candida tropicalis |
60 |
Fluconazole |
1 |
S |
|
Voriconazole |
≤0.12 |
S |
||
|
Caspofungin |
≤0.12 |
- |
||
|
Micafungin |
≤0.06 |
- |
||
|
Amphotericin B |
≤0.25 |
S |
||
|
127 |
Fluconazole |
≤0.5 |
S |
|
|
Voriconazole |
≤0.12 |
S |
||
|
Caspofungin |
≤0.12 |
- |
||
|
Micafungin |
≤0.06 |
- |
||
|
Amphotericin B |
≤0.25 |
S |
MIC: Minimum Inhibitory Concentration. EUCAST v.10.0. S: Sensible. R: Resistant. I: Susceptible, increased exposure. IE: indicates that there is insufficient evidence that the species in question is a good target for therapy with the drug.
Multi-Locus Sequence Typing
Molecular epidemiology analysis based on MLST genes forC. albicansidentified three new sequence types (STs): ST4220, ST4278, and ST4279. Additionally, a new allele for the AAT1a gene (allele 215) was registered in the PubMLST database, originating from sample 126. ForC. krusei, a new sequence type, ST303, was identified as circulating in the hospital during the study period. In contrast,C. tropicalisisolates were classified as ST1248 and ST327, previously reported in China and Taiwan, respectively, and identified from blood samples (Table 3).
Table 3: Types of STs identified in Candida species isolated from oncohematological patients from Hemoam Foundation
|
Nº |
Species |
ST |
Continents/Regions |
Sample |
Comments |
|
1 |
C. albicans |
691 |
Africa, Asia, Australia, China, United Kingdom, United States of America, Brazil, France, Portugal, Austaly, New Zlandia, Norway, Ireland, Japan, Morocco, |
feces, oropharynx, skin, blood, swab and urine, sputum |
common ancestral of all ST |
|
4431 |
China |
Blood, urine, oropharynx, vaginal swab |
most recent common ancestor of ST2904 |
||
|
4431 |
Taiwan |
blood |
most recent common ancestor of ST42792 |
||
|
791 |
China, United States, United Kingdom, Kuwait, Argentina, Colombia, Argentina, France, Japan, Portugal, Morocco, Portugal |
urine, vaginal swab, blood, sputum |
most recent common ancestor of ST42202 |
||
|
17541 |
China |
oropharynx |
most recent common ancestor of ST90 |
||
|
901 |
South Africa, Kuwait, South Korea, Brazil, Guyana, China |
blood, feces, oropharynx, urine, vaginal swab |
most recent common ancestor of ST42782 |
||
|
42202 |
Brazil |
blood, urine |
- |
||
|
42782 |
Brazil |
blood |
- |
||
|
42792 |
Brazil |
blood |
- |
||
|
2641 |
China |
Blood, other sterile sites |
Common ancestral of all ST |
||
|
2 |
C. krusei |
19 |
United Kingdom, Canada, Australia |
not informed |
most recent common ancestor of ST24 |
|
241 |
United Kingdom, Poland, Taiwan, France |
blood |
most recent common ancestor of ST69 |
||
|
691 |
Czech |
blood |
common ancestor of ST3032 |
||
|
3032 |
Brazil |
blood |
- |
1: PubMLST data. 2: New ST (this study)
e-BURST analysis ofC. albicanssequence types showed that ST69 is the founding clone from which all the newly identified STs likely originated. This clone has been found in multiple regions and countries, as well as in various biological sources, including feces, oropharyngeal swabs, skin, blood, vaginal swabs, sputum, and urine (Table 2). ST4220 and ST4279 seem to cluster within the ST69 clonal complex and are part of two separate subgroups formed by ST443, ST2904, and ST76. Both ST4220 and ST4279 are located as terminal nodes, indicating recent divergence and potential for future subgroup development. In contrast, ST4278 is part of a subgroup that traces back to ST1754 and shares ST90 as a common ancestor, also forming a distinct terminal node (Supplemental material- S1).
S1. Evolutionary relationship of C. albicans STs isolated from oncohematological patients at the FHEMOAM in Manaus, Brazil.
RegardingC. krusei, the eBURST diagram (Figure 2) identified ST256, initially reported in China, as the ancestral clone. The new ST303 appears to have descended from ST69, a clone previously isolated from blood samples in the Czech Republic. Both are part of a clonal complex derived from ST19, a sequence type that circulates in the United Kingdom, Canada, Australia, and has also been detected in animal hosts [17] (Supplemental material- S2).
S2. Evolutionary relationship of C. krusei ST isolated from an oncohematological patient at the FHEMOAM in Manaus, Brazil.
Discussion
Invasive fungal infections (IFIs) continue to pose a major challenge in hematology, withCandida species being the primary pathogens in this setting [29]. Candidiasis is among the most common invasive fungal diseases linked to bloodstream spread, especially in patients with hematological cancers, such as those treated at FHEMOAM [1,29].
In this series, three notable species—C. albicans,C. tropicalis, andC. krusei—were isolated from blood and urinary infections in hospitalized patients with oncohematological conditions. Notably,C. albicansandC. tropicalisare designated as WHO-priority fungal pathogens [16]. Similar findings have been reported by Chen et al. (China), Mitra et al. (India), Vargas-Espindola et al. (Colombia), and Diniz et al. (Brazil), all of whom identified Candida species associated with blood and urinary tract infections in neutropenic or immunocompromised patients [6,9,25,29].
Beyond the serious nature of these infections and the risks of morbidity and death, the circulating resistance of C. krusei to amphotericin B and fluconazole at FHEMOAM raises concerns and requires attention. While ourC. tropicalisisolates were fully susceptible, previous studies in Thailand and Colombia reported fluconazole-resistantC. tropicalisstrains [6,8]. In Brazil, Diniz et al. [25] noted thatC. albicansandC. tropicalisremained sensitive to amphotericin B, fluconazole, and echinocandins, aligning with our results. These differences likely reflect variation in antifungal use and local environmental pressures influencing susceptibility patterns. Of particular interest is the resistance profile ofC. krusei, which has been documented across Asia, Europe, North America, and South America. Our findings agree with those of Domán et al. [17], who identifiedC. kruseistrains with inherent fluconazole resistance or acquired amphotericin B resistance, often linked to mutations in the hotspot regions (HS1, HS2, HS3) of the conservedFKS1gene, which encodes the target of echinocandins, underscoring the importance of ongoing antifungal monitoring [26].
Our molecular epidemiology results also reveal the genetic diversity and worldwide distribution ofCandidaclones. The ancestry analysis of new C. albicans sequence types (ST4220 and ST4279) showed that ST69, previously identified by Tian et al. [27] and McManus et al. [18], is a dominant global founder genotype. This clone is highly capable of colonizing humans and has been isolated from various anatomical sites and regions [18,28]. Although ST4278 is a novel C. albicans sequence type from our study, it shows a distinct evolutionary pathway with ST90 as it’ s closest ancestor. Despite being isolated from patients in the same facility, the threeC. albicans types (ST4220, ST4279, and ST4278) occupy different branches in the eBURST diagram, indicating significant genomic variation. Similar genetic diversity has been observed in Kuwait, Germany, Taiwan, and Singapore, reflecting the complex population structure ofC. albicans [18,27,29]. To our knowledge, this is thefirst report of novel STs ofC. albicansandC. kruseicirculating in a Hematology Reference Center in Northern Brazil. The combination of antifungal susceptibility profiles and molecular typing underscores the importance of ongoing surveillance in immunocompromised populations [7,30]. Antifungal prophylaxis, when used judiciously, may help decrease morbidity and mortality in this setting [30,31].
Conclusion
This study identified new Candida STs 4220, 4279, and 4278 (C. albicans) and ST303 (C. krusei) at a Hematologic Reference Center in Brazil. These results highlight the circulation of previously unreported STs in this region. Amphotericin B- and fluconazole-resistant C. krusei is especially concerning due to the limited antifungal options for hematologic patients. Possibly influenced by local selective pressure, such as antifungal therapy, microevolutionary processes may promote the emergence of these resistant clones. While some of these STs share ancestry with globally widespread genotypes like ST69, their different evolutionary paths could support the idea of local adaptation and potential clonal spread within a hospital setting. Ongoing molecular surveillance, along with prudent antifungal use and informed treatment choices, may be importante for monitoring clonal spread and guiding infection control strategies, as well as preventing outbreaks. These findings support the hypothesis that a new Candida ST maybe present in our hospital environment.
Acknowledgements
The authors are grateful to Amazonas State Research Support Foundation (FAPEAM), Brazil, for granting a scientific initiation scholarship, to João Paulo Diniz Pimentel and Miriam Rodrigues Ribeiro Santiago.
Declarations
Ethics statement
This study was approved by the Foundation Human Research Ethical Committee (CEP/HEMOAM) under CAAE Nº 68471223.5.0000.0009.
Competing interests
The authors declare no competing interests.
Author’s Contributions
CMF, WAF co-conceptualized the study, undertook statistical analyses and wrote the original manuscript draft. CMF, WAF, FGN, GAVS, AMT, GMAF, JSC revised the mansucript.
GMAF, LAG, ARP, JSC, AMT and FOC undertook sample collection and laboratory tests. VCS, FGN AND GAVS sequenced the isolates. CMF, WAF and GMAF supervised the work, vetted the results and critically reviewed the manuscript.
References
- Camplesi Junior M, Silva HM, Arantes AM, et al. Invasive fungal infection in patients with hematologic disorders in a Brazilian tertiary care hospital. Rev Soc Bras Med Trop 50 (2017): 80–5.
- Da Matta DA, Melo AS, Guimarães T, et al. Multilocus sequence typing of sequential Candida albicans Iso/lates from patients with persistent or recurrent Fungemia. Medical Mycology 48 (2010): 757–762.
- Afsarian SMH, Badali H, Shokohi T, Najafipour S. Molecular Diversity of Candida albicans Isolated from Immunocompromised Patients, Based on MLST Method. Iran J Public Health 44 (2015): 1262-1269.
- Sanguinetti M, Posteraro B, Lass-Florl C. Antifungal drug resistance among Candida species: mechanisms and clinical impact. Mycoses 58 (2015): 2–13.
- Knoll MA, Lackner N, Ulmer H, et al. Multiple-antifungal colony susceptibility testing detects polyresistance in clinical Candida cultures: a European Confederation of Medical Mycology excellence center study. Clinical Microbiology and Infection 28 (2022): 1288.
- Bergamasco MD, Garnica M, Colombo AL, Nucci M. Epidemiology of candidemia in patients with hematologic malignancies and solid tumours in Brazil. Mycoses 56 (2012): 256-263.
- Vargas-Espíndola LA, Cuervo-Maldonado SI, Enciso-Olivera JL, et al. Fungemia in Hospitalized Adult Patients with Hematological Malignancies: Epidemiology and Risk Factors. J Fungi 9 (2023): 400.
- Shafiee F, Soltani R, Meidani M. Invasive fungal infections in hematologic malignancies: Incidence, management, and antifungal therapy. J Res Med Sci 28 (2023): 73.
- Boonsilp S, Homkaew A, Phumisantiphong U, Nutalai D. Species Distribution, Antifungal Susceptibility and Molecular Epidemiology of Candida Species Causing Candidemia in a Tertiary Care Hospital in Bangkok, Thailand. Journal of Fungi 7 (2021): 577.
- Mitra AN, Pramanik P, Bhattacharya R. Invasive Fungal Infections in Acute Haematological Malignancies: A Cross-sectional Study. Journal of Clinical and Diagnostic Research 17 (2023): DC07-DC13.
- Cornely OA, Gachot B, Akan H, et al. Epidemiology and Outcome of Fungemia in a Cancer Cohort of the Infectious Diseases Group (IDG) of the European Organization for Research and Treatment of Cancer (EORTC 65031). Clin. Infect. Dis 61 (2015): 324–331.
- Sipsas NV, Lewis RE, Tarrand J, et al. Candidemia in Patients with Hematologic Malignancies in the Era of New Antifungal Agents (2001-2007): Stable Incidence but Changing Epidemiology of a Still Frequently Lethal Infection. Cancer 115 (2009): 4745–4752.
- Zirkel J, Klinker H, Kuhn A, et al. Epidemiology of Candida Blood Stream Infections in Patients with Hematological Malignancies or Solid Tumors. Mycol 50 (2012): 50-55.
- Silva LM, Ferreira WA, Filho RAAB, et al. New ST623 of Cryptococcus neoformans isolated from a patient with non-Hodgkin’s lymphoma in the Brazilian Amazon. Ann Clin Microbiol Antimicrob 20 (2020).
- Ramírez I, Moncada D. Fatal Disseminated Infection by Trichosporon asahii Under Voriconazole Therapy in a Patient with Acute Myeloid Leukemia: A Review of Breakthrough Infections by Trichosporon spp. 1o de abril de 185 (2020): 377–88.
- León-Buitimea A, Garza-Cervantes JA, Gallegos-Alvarado DY, et al. Nanomaterial-based antifungal therapies to combat fungal diseases aspergillosis, coccidioidomycosis, mucormycosis, and candidiasis. Pathogens 10 (2021).
- WHO fungal priority pathogens list to guide research, development, and public health action. Geneva: World Health Organization (2022).
- Domán M, Makrai L, Bányai K. Molecular Phylogenetic Analysis of Candida krusei. Mycopathologia 187 (2022): 333–343.
- McManus B, Coleman DC. Molecular epidemiology, phylogeny and evolution of Candida albicans. Infection, Genetics and Evolution 21 (2014): 166–178.
- Bougnoux ME, Tavanti A, Bouchier C,et al. Collaborative Consensus for Optimized Multilocus Sequence Typing of Candida albicans. Journal of Clinical Microbiology 41 (2003): 5265–5266.
- Tavanti A, Davidson AD, Johnson EM, et al. Multilocus Sequence Typing for Differentiation of Strains of Candida tropicalis. Journal of Clinical Microbiology 43 (2005): 5593–5600.
- Jacobsen MD, Gow NAR, Maiden MCJ, et al. Strain Typing and Determination of Population Structure of Candida krusei by Multilocus Sequence Typing. Journal of Clinical Microbiology 45 (2007): 317–323.
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Research 3 (2018): 124.
- Feil EJ, Li BC, Aanensen DM,et al. eBURST: Inferring Patterns of Evolutionary Descent among Clusters of Related Bacterial Genotypes from Multilocus Sequence Typing Data. Journal of Bacteriology 186 (2004): 1518–1530.
- Francisco AP, Bugalho M, Ramirez M, and Carriço JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10 (2009): 152.
- Diniz MV, Araújo PSR, Barbosa RNF, et al. Bloodstream infection by Candida in patients with hematologic neoplasia: polyphasic taxonomy and antifungal susceptibility. Brazilian Journal of Health Review 5 (2022): 3774-3786.
- Forastiero A, Garcia-Gil V, Rivero-Menendez O, et al. Rapid development of Candida krusei echinocandin resistance during caspofungin therapy. Antimicrob Agents Chemother 59 (2015): 6975–6982.
- Tian JY, Yang Y, Chen S, et al. Genetic diversity, and molecular epidemiology of Candida albicans from vulvovaginal candidiasis patients. Infection, Genetics and Evolution 92 (2021): 104893.
- Ge SH, Xie J, Xu J, et al. Prevalence of specific and phylogenetically closely related genotypes in the population of Candida albicans associated with genital candidiasis in China. Fungal Genetics and Biology 49 (2012): 86–93.
- Chew KL, Achik R, Osman NH, et al. Genomic epidemiology of human candidaemia isolates in a tertiary hospital. Microbial Genomics 9 (2023): 001047.
- Bandeira LRB, Cristina JS, Mendes AGR, et al. Use of antifungal prophylaxis in pediatric patients undergoing leukemia treatment at a reference center in Brazilian Amazon. Peer Review 6 (2024): 346-359.
- Fracchiolla NS, Sciumè M, Orofino N, et al. Epidemiology and treatment approaches in management of invasive fungal infections in hematological malignancies: Results from a single-Centre study. PLoS One 14 (2019): e0216715.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
