Uncovering Carbapenem Resistance: A Molecular Look at Klebsiella pneumoniae in Clinical Samples
Dr. Nooriya Haque*,1, Prof. Dr. SM Shamsuzzaman2, Dr. Tarafder Mohammad Atiquzzaman3, Dr. Rafia Afreen Jalil4, Dr. Shaila Akhtar5, Dr. Md. Izaz Miah6, Dr. Md. Sirazum Munir7
1Assistant Professor (C.C), Department of Microbiology, Green Life Medical College, Dhaka, Bangladesh
2Professor, Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh
3Medical Officer, Department of Paediatric Surgery, Bangladesh Medical University, Dhaka, Bangladesh
4Associate Professor, Department of Microbiology, Green Life Medical College, Dhaka, Bangladesh
5Assistant Professor (C.C), Department of Microbiology, Green Life Medical College, Dhaka, Bangladesh
6Lecturer, Department of Microbiology, Manikganj Medical College, Manikganj, Bangladesh
7Anesthesiologist, Intensive Care Unit Laboratory, Rajshahi Medical College Hospital, Rajshahi, Bangladesh
*Corresponding author: Dr. Nooriya Haque, Assistant Professor (C.C), Department of Microbiology, Green Life Medical College, Dhaka, Bangladesh.
Received: 12 July 2025; Accepted: 17 July 2025; Published: 29 July 2025
Article Information
Citation: Dr. Nooriya Haque, Prof. Dr. SM Shamsuzzaman, Dr. Tarafder Mohammad Atiquzzaman, Dr. Rafia Afreen Jalil, Dr. Shaila Akhtar, Dr. Md. Izaz Miah, Dr. Md. Sirazum Munir. Uncovering Carbapenem Resistance: A Molecular Look at Klebsiella pneumoniae in Clinical Samples. Archives of Microbiology and Immunology. 9 (2025): 223-229.
View / Download Pdf Share at FacebookAbstract
The rise of multidrug-resistant (MDR) Klebsiella pneumoniae has greatly complicated the management of infections caused by this pathogen. This study aimed to assess the antimicrobial resistance patterns and to detect the presence of β-lactamase genes in both MDR and non-MDR K. pneumoniae isolates obtained from a variety of clinical specimens. A total of 50 K. pneumoniae isolates were collected and identified using standard microbiological techniques. Antimicrobial susceptibility testing was performed using the Kirby-Bauer disk diffusion method. Phenotypic detection of extended-spectrum β-lactamases (ESBLs), metallo-β-lactamases (MBLs), and carbapenemases was carried out via the double-disk synergy test, combined disk test, and modified Hodge test, respectively. Detection of resistance and virulence genes at the molecular level was conducted using polymerase chain reaction (PCR). Multidrug resistance was observed in 50% of the isolates, with high levels of resistance to β-lactam antibiotics, carbapenems, ciprofloxacin, piperacillin, and tazobactam. Notably, 28% of the isolates also exhibited resistance to colistin. ESBL was identified phenotypically in 30% of the isolates. NDM-1 (12%), NDM-5 (2%), VIM (4%), and KPC (8%) were detected among β-lactamase genes. These findings indicate a high prevalence of MDR K. pneumoniae in Bangladesh, posing a serious challenge for infection control and treatment strategies in healthcare settings.
Keywords
<p>ESBL; Klebsiella pneumoniae; MDR; Bangladesh; Antibiotic resistance</p>
Article Details
Introduction
Carbapenems are a group of broad-spectrum β-lactam antibiotics commonly prescribed as a last resort for treating infections caused by multidrug-resistant Gram-negative bacteria. However, the emergence of carbapenem-resistant Klebsiella pneumoniae (CR-Kp) has become a major concern in hospitals worldwide due to their link to severe infections and high morbidity and mortality rates (1). Clinical isolates of K. pneumoniae are known for producing a range of β-lactamase enzymes and are intrinsically resistant to certain antibiotics such as ampicillin and amoxicillin (2). The increasing frequency and rapid spread of resistant strains present a growing clinical challenge. Resistance to β-lactam antibiotics in this species is primarily due to the production of enzymes like extended-spectrum β-lactamases (ESBLs), plasmid-mediated AmpC β-lactamases, and carbapenemases. CR-Kp isolates often exhibit resistance not only to carbapenems but also to multiple other antibiotic classes, including penicillins, third-generation cephalosporins, fluoroquinolones, and aminoglycosides. The prevalence of these resistant strains varies by region and is influenced by local infection control practices and antimicrobial stewardship efforts (3). Of particular concern are emerging pan-resistant strains of CR-Kp, which leave no effective antibiotic options and pose a serious global health threat. Understanding the molecular epidemiology and resistance mechanisms of these strains is critical for identifying potential alternative treatments and controlling their spread (4).
One of the primary mechanisms of carbapenem resistance in K. pneumoniae involves the production of carbapenemase enzymes (5). These enzymes are categorized into four Ambler classes—A, B, C, and D—based on their molecular structure. Among them, classes A (e.g., KPC), B (e.g., NDM, VIM, IMP), and D (e.g., OXA-48-like enzymes) are particularly relevant in clinical settings (6). It is not uncommon for a single strain to harbor multiple β-lactamase genes, which may contribute to its high adaptability and resistance profile (7). These genes are often co-located on mobile genetic elements like plasmids and transposons, facilitating their transfer and persistence across bacterial populations (5). In this study, we sought to characterize the β-lactamase genes present in carbapenem-resistant K. pneumoniae isolates collected from clinical samples in Bangladesh. Understanding these genetic resistance patterns is essential for informing treatment strategies and for curbing the spread of carbapenem-resistant Gram-negative bacteria.
Materials and Methods
Study Design and Setting
A cross-sectional study was carried out in the Department of Microbiology and Immunology at Dhaka Medical College Hospital (DMCH), Dhaka, Bangladesh. The research was conducted over one year, from January to December 2022.
Isolation and Identification of Klebsiella pneumoniae
Clinical specimens, including wound swabs, urine, wound swabs, pus, tracheal aspirates, sputum, blood, and other body fluids, submitted to the Microbiology Laboratory at DMCH were processed for bacterial isolation. A total of 50 consecutive, non-duplicate isolates of K. pneumoniae were obtained from hospitalized patients during the study period. Identification of K. pneumoniae began with observing colony morphology on MacConkey agar, followed by Gram staining and standard biochemical tests. These included catalase and oxidase activity, urease production, indole reaction, gas formation, motility testing, citrate utilization, and lactose fermentation. For quality control, the reference strain K. pneumoniae ATCC 700603 was used during culture, biochemical testing, and phenotypic confirmation of clinical isolates. The study received ethical approval from the institutional review board of Dhaka Medical Colleg, and written informed consent was secured from all participating patients.
Antimicrobial Susceptibility Testing
The antimicrobial susceptibility profiles of the Klebsiella pneumoniae isolates were determined using the standard Kirby-Bauer disk diffusion method on Mueller-Hinton agar (Oxoid Ltd., UK) (8). A range of commercially prepared antibiotic discs was employed, including amoxicillin-clavulanic acid (20/10 µg), piperacillin-tazobactam (100/10 µg), cefepime (30 µg), cefoxitin (30 µg), ceftriaxone (30 µg), cefuroxime (30 µg), ceftazidime (30 µg), amikacin (30 µg), ciprofloxacin (5 µg), imipenem (10 µg), tigecycline (15 µg), and aztreonam (30 µg). Interpretation of inhibition zones was carried out under guidelines established by the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST). To identify ESBL-producing strains, the double-disk synergy test was applied. Furthermore, isolates were categorized as multidrug-resistant (MDR), extensively drug-resistant (XDR), or pandrug-resistant (PDR) based on criteria outlined by the Centers for Disease Control and Prevention (CDC) (9, 10).
Definition of Multidrug-Resistant (MDR) Klebsiella pneumoniae
In this study, Klebsiella pneumoniae isolates were classified as multidrug-resistant (MDR) if they demonstrated resistance to at least one antimicrobial agent in three or more distinct classes of antibiotics. These classes included, but were not limited to, antipseudomonal penicillins, aminoglycosides, carbapenems, first and second-generation cephalosporins, extended-spectrum cephalosporins, third and fourth-generation cephalosporins, fluoroquinolones, β-lactam/β-lactamase inhibitor combinations, and penicillins, following CDC-defined criteria (10).
Phenotypic Detection of Resistance Mechanisms
Detection of ESBL Production Using the Double Disk Synergy Test (DDST)
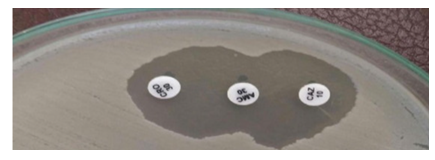
ESBL activity was evaluated phenotypically using the double disk synergy test (DDST) on Mueller-Hinton Agar (MHA). In this method, disks containing third-generation cephalosporins, specifically ceftriaxone and ceftazidime (30 µg each), were positioned 20 mm apart from a central disk of amoxicillin-clavulanic acid (20/10 µg). An enhancement of the inhibition zone around either cephalosporin disk in relation to the clavulanate-containing disk was interpreted as an ESBL production. A distinct "keyhole" or "champagne cork" shaped zone confirmed the presence of synergy between the antibiotics, supporting the ESBL positive phenotype (10) (Fig. 1).
Detection of Carbapenemase Production Using the Modified Hodge Test (MHT)
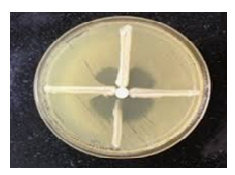
The Modified Hodge Test (MHT) was employed to screen for carbapenemase activity in the Klebsiella pneumoniae isolates. A suspension of Escherichia coli ATCC 25922 was prepared and adjusted to a 0.5 McFarland turbidity standard. Using a sterile cotton swab, the suspension was uniformly spread over the surface of a Mueller-Hinton agar plate to form a lawn culture. The inoculated plate was then left at room temperature for approximately 15 minutes to allow for surface drying. An imipenem (10 µg) disk was placed at the center of the agar plate. Test isolates were streaked in a straight line from the edge of the imipenem disk outward toward the edge of the plate. After overnight incubation at 37°C, plates were examined for enhanced growth of E. coli along the streaked line toward the imipenem disk. A cloverleaf-like indentation in the inhibition zone around the disk at the point of intersection was considered a positive result, indicating carbapenemase production by the test organism (11) (Fig. 2).
Combined Disc Test with Imipenem-EDTA for Detection of Metallo-β-Lactamase (MBL)
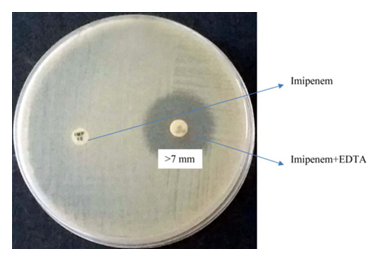
To screen for the presence of metallo-β-lactamase (MBL) production, the combined disc test using imipenem and imipenem-EDTA was performed. The test isolate was adjusted to a 0.5 McFarland turbidity standard and uniformly inoculated onto Mueller-Hinton agar using a sterile swab. Two antibiotic discs were placed on the agar surface: one containing imipenem (10 µg) and the other containing imipenem combined with EDTA (10/750 µg). The plates were incubated at 37°C for 18–24 hours. An increase of ≥7 mm in the diameter of the inhibition zone surrounding the imipenem-EDTA disc compared to the imipenem-only disc was interpreted as indicative of MBL production by the isolate (12, 13) (Fig. 3).
Detection of β-Lactamase Genes by Polymerase Chain Reaction (PCR)
Conventional polymerase chain reaction (PCR) was employed to detect carbapenemase (KPC) and metallo-β-lactamase (NDM, VIM) genes in Klebsiella pneumoniae isolates. Genomic DNA was extracted using the boiling method. Amplified PCR products were visualized by agarose gel electrophoresis to confirm the presence of target genes (14,15).
Data Analysis
All collected data were analyzed using IBM SPSS Statistics version 23. Categorical variables were summarized as frequencies and percentages. Graphs and visualizations were generated using the matplotlib library in Python.
Results
Out of a total of 350 clinical specimens analyzed, the majority were wound swabs and pus (n = 149), followed by urine samples (n = 91), sputum (n = 42), blood (n = 46), and endotracheal aspirates (n = 22). Of these, 226 samples (64.57%) showed positive bacterial growth upon culture, as detailed in Table 1. The distribution of bacterial species identified from these 226 culture-positive samples is presented in Table 2.
Table 1: Positive culture results from various clinical samples (N = 350).
|
Samples |
Number of Samples |
Culture Positive n (%) |
|
Wound swab and pus |
149 |
115 (77.18) |
|
Urine |
91 |
45 (49.45) |
|
Sputum |
42 |
23 (54.76) |
|
ETA |
22 |
19 (86.36) |
|
Blood |
46 |
24 (52.17) |
|
Total |
350 |
226 (64.57) |
Table 2: The distribution of organisms isolated from various samples (n = 226).
|
Organism |
n (%) |
|
Escherichia coli |
54 (23.89) |
|
Pseudomonas aeruginosa |
51 (22.57) |
|
Klebsiella pneumoniae |
50 (22.12) |
|
Klebsiella oxytoca |
2 (0.88) |
|
Acinetobacter baumanii |
13 (5.75) |
|
Enterobacter cloacae |
8 (3.54) |
|
Enterobacter aerogenes |
6 (2.65) |
|
Citrobacter freundii |
2 (0.88) |
|
Citrobacter koseri |
1 (0.44) |
|
Proteus mirabilis |
5 (2.22) |
|
Proteus vulgaris |
3 (1.34) |
|
Gram-positive bacteria |
31 (13.72) |
|
Total |
226 (100.00) |
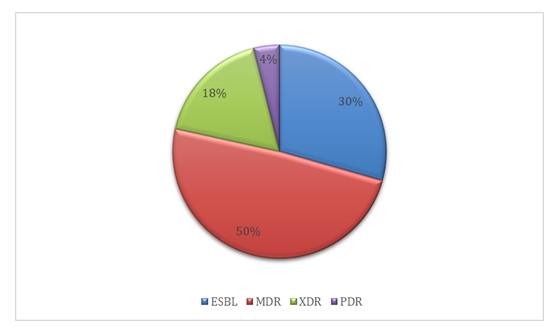
The antimicrobial resistance profiles of the Klebsiella pneumoniae isolates are summarized in Table 3. Of the 50 isolates tested, resistance to cefuroxime was observed in 66% (n = 33), followed closely by ceftriaxone, with 62% (n = 32) of isolates showing resistance. Fosfomycin exhibited the highest susceptibility among the tested antibiotics, with only 12% (n = 6) of isolates demonstrating resistance. Notably, resistance to colistin was identified in 28% (n = 14) of the isolates. The dissemination of resistance categories amid the isolates is presented in Table 4. Half of the K. pneumoniae isolates (n = 25, 50%) were classified as multidrug-resistant (MDR), while 18% (n = 9) met the criteria for extensively drug resistant (XDR) status. Two isolates (4%) were identified as pandrug resistant (PDR), and 15 isolates (30%) were confirmed as ESBL producers, as illustrated in Figure 4.
Table 3: Antimicrobial resistance pattern among isolated Klebsiella pneumoniae (n=50)
|
Antimicrobial drugs |
Resistance, n (%) |
|
Amikacin |
23 (46.0) |
|
Amoxiclav |
24 (48.0) |
|
Cefoxitin |
24 (48.0) |
|
Cefuroxime |
33 (66.0) |
|
Ceftazidime |
30 (60.0) |
|
Ceftriaxone |
32 (64.0) |
|
Cefepime |
25 (50.0) |
|
Ciprofloxacin |
26 (52.0) |
|
Piperacillin/Tazobactam |
18 (36.0) |
|
Aztreonam |
31 (62.0) |
|
Imipenem |
11 (22.0) |
|
Tigecycline |
9 (18.0) |
|
Fosfomycin |
6 (12.0) |
|
Colistin |
14 (28.0) |
Table 4: Classification of Klebsiella pneumoniae isolates by resistance profile, including MDR, XDR, PDR, and ESBL producers (n = 50)
|
Type of resistance |
n (%) |
|
ESBL |
15(30.0) |
|
MDR |
25(50.0) |
|
XDR |
9(18.0) |
|
PDR |
2 (4.0) |
Phenotypic detection of resistance mechanisms among Klebsiella pneumoniae isolates is summarized in Table 5. Of the 50 isolates examined, extended-spectrum β-lactamase (ESBL) production was identified in 15 isolates (30.0%) using the double disc synergy test (DDST). Metallo-β-lactamase (MBL) activity was detected in 11 isolates (22.0%) through the combined disc (CD) test with imipenem and EDTA. Additionally, carbapenemase production was confirmed in 8 isolates (16.0%) using the Modified Hodge Test (MHT).
Table 5: Phenotypic detection of β-Lactamase production in Klebsiella pneumoniae isolates (n = 50)
|
Detection Method |
Positive Isolates, n (%) |
|
Double Disc Synergy Test (ESBL) |
15 (30.0%) |
|
Combined Disc Assay (MBL) |
11 (22.0%) |
|
Modified Hodge Test (Carbapenemase) |
8 (16.0%) |
Among the 50 Klebsiella pneumoniae isolates analyzed, metallo-β-lactamase (MBL) genes were detected in 11 isolates (22.0%). Of these, six isolates (12.0%) carried the NDM-1gene, one isolate (2.0%) harbored NDM-5, and two isolates (4.0%) tested positive for the VIM gene. In addition, carbapenemase genes were identified in 8 isolates (16.0%), with KPC detected in four of them (8.0%), as detailed in Table 6.
Table 6: Distribution of MBL and carbapenemase genes detected in Klebsiella pneumoniae isolates (n = 50)
|
Gene Type |
Gene |
Positive Isolates, n |
Percentage (%) |
|
Metallo-β-lactamase (MBL) |
NDM-1 |
6 |
12 |
|
|
NDM-5 |
1 |
2 |
|
VIM |
2 |
4 |
|
|
Total (MBL) |
9 |
18 |
|
|
Carbapenemase |
KPC |
4 |
8 |
|
Total |
4 |
8 |
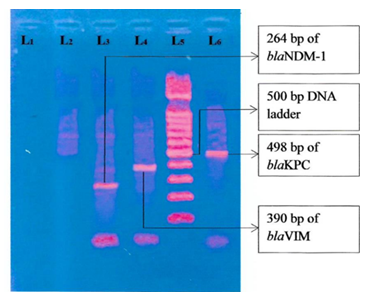
Figure 5: Photograph of gel electrophoresis; negative control without DNA (TE buffer) (lane one), negative control Escherichia coli ATCC 25922 (lane 2), amplified DNA of 264 bp for blaNDM-1 (lane-3), DNA of 390 bp of blaVIM (lane 4), DNA of 498 bp of blaKPC (lane 6) in imipenem-resistant K. pneumoniae, hundred bp DNA ladder (lane 5).
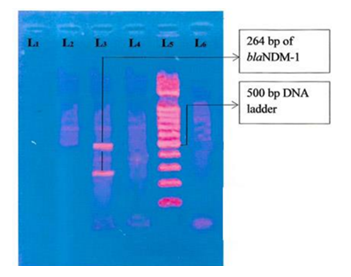
Figure 6: Photograph of gel electrophoresis; negative control without DNA (TE buffer) (lane one), negative control Escherichia coli ATCC 25922 (lane 2), amplified DNA of 264 bp for blaNDM-1(lane-3), DNA of 498 bp of blaKPC (lane 6) in imipenem-resistant K. pneumoniae, hundred bp DNA ladder (lane 5).
Discussion
In this study, 226 out of 350 non-duplicate clinical specimens (64.57%) showed positive bacterial growth. Among the culture-positive cases, Escherichia coli was the most frequently isolated organism, accounting for 23.89%, followed by Pseudomonas aeruginosa (22.57%) and Klebsiella pneumoniae (22.12%). This distribution aligns with similar findings from a study in India, where 56.9% of clinical samples yielded bacterial growth (16). The prevalence of K. pneumoniae observed in this study is consistent with earlier reports from Bangladesh, which documented rates of 24% in the northeastern region and 19.72% in the southeast (17,18). Among the 50 K. pneumoniae isolates analyzed, half (n = 25; 50%) were identified as multidrug-resistant (MDR). This proportion is comparable to findings from a study in Indonesia by Nirwati et al., where 54.49% of isolates were MDR (19). However, a considerably higher MDR prevalence (84.37%) was reported in Iraq by Aljanaby et al. (20), indicating geographical variability in resistance patterns. The resistance profile of the isolates showed elevated resistance to second through fourth-generation cephalosporins, aztreonam, and ciprofloxacin. Similar resistance trends were previously reported in Bangladesh, where K. pneumoniae isolates displayed high resistance to cefuroxime, ceftriaxone, ceftazidime, cefepime, and ciprofloxacin (21,22).
In our study, the rates of resistance were notable: cefuroxime (66%), ceftriaxone (64%), aztreonam (62%), ceftazidime (60%), ciprofloxacin (52%), and cefepime (50%). These findings are in line with results from Kawser et al., who reported comparable resistance rates (23). Resistance to last-resort antibiotics, including colistin (28%), imipenem (22%), tigecycline (18%), and fosfomycin (12%), was also observed. Colistin resistance, in particular, has shown an upward trend in Bangladesh, with a previous systematic review reporting median resistance rates of 18.8%, ranging from 0% to 21.4% (22,24). This increase is likely influenced by the clinical overuse of colistin and horizontal transfer of resistance genes via mobile genetic elements. Phenotypic screening for extended-spectrum β-lactamase (ESBL) production using the double-disc synergy test (DDST) identified 15 isolates (30%) as ESBL producers. This result aligns with a study by Chakraborty et al. in Bangladesh, which reported a 45% ESBL rate in K. pneumoniae isolates(25), while a lower rate of 17% was observed in a study from Pakistan by Riaz et al (26). Metallo-β-lactamase (MBL) production was detected in 11 isolates (22%) through the combined disc assay. This is notably lower than the 85.7% MBL detection rate among imipenem-resistant isolates reported by Farzana et al (27). The discrepancy may be due to the inclusion of all K. pneumoniae isolates in the current study, rather than only imipenem non-susceptible ones, which may have lowered the overall detection rate.
Carbapenemase activity was identified in 8 isolates (16%) using the Modified Hodge Test (MHT). This detection rate is lower than those reported in studies by Metwally and Eftekhar, which found carbapenemase production in 70% and 90% of isolates, respectively (28,29). The sensitivity and specificity of the MHT can vary by region and testing conditions, and its limitations have been noted in previous literature, including by Nordmann et al (30). Molecular analysis revealed that 22% of the isolates carried MBL genes. Specifically, NDM-1 was detected in 12% of isolates, and NDM-5 in 2%. These findings are consistent with previous studies conducted in Bangladesh, where Farzana et al. and Rahman et al. reported NDM gene presence in 22.86% and 20.51% of isolates, respectively (31,32). The VIM gene was found in 4% of isolates. As previously described by Queenan et al., VIM and IMP genes are more frequently associated with P. aeruginosa and are less commonly detected in Enterobacteriaceae (33). Carbapenemase gene detection revealed that 8 isolates (16%) harbored such genes, with KPC identified in 4 isolates (8%). This is somewhat lower than the 25% prevalence reported by Sattar et al. in a study of K. pneumoniae isolates (34).
Conclusion
Most of the Klebsiella pneumoniae isolates were multidrug-resistant in this study. There are limited therapeutic options to treat MDR Klebsiella pneumoniae, and this manifests a gloomy scenario for the management of hospital infections in Bangladesh; it further bears the necessity of necessary surveillance system to deal with the inevitable healthcare disaster in the offing.
Ethical Clearance
Ethical approval for this study was granted by the Institutional Review Board.
Financial Support and Sponsorship
None.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Almaghrabi R, Clancy CJ, Doi Y, et al. Carbapenem-Resistant Klebsiella pneumoniae Strains Exhibit Diversity in Aminoglycoside-Modifying Enzymes, Which Exert Differing Effects on Plazomicin and Other Agents. Antimicrob Agents Chemother 58 (2014): 4443–51.
- Li J, Shi Y, Song X, et al. Mechanisms of Antimicrobial Resistance in Klebsiella: Advances in Detection Methods and Clinical Implications. IDR 18 (2025): 1339–54.
- Hou X hua, Song X yu, Ma X bo, et al. Molecular characterization of multidrug-resistant Klebsiella pneumoniae isolates. Braz J Microbiol 46 (2015): 759–68.
- Wernli D, Haustein T, Conly J, et al. A Call for Action: The Application of the International Health Regulations to the Global Threat of Antimicrobial Resistance. PLoS Med 8 (2011): e1001022.
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiology Reviews 41 (2017): 252–75.
- Schwabbauer ML. Use of the latent image technique to develop and evaluate problem-solving skills. Am J Med Technol 41 (1975): 457–62.
- Lee CR, Lee JH, Park KS, et al. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front Microbiol 13 (2016): 7.
- AW Bauer, WMM Kirby, JC Sheris, et al. Antibiotic susceptibility testing by a standardized single disk method Am. J. clin. pathol 45 (1966): 493-496.
- Wayne Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: 20th informational supplement CLSI document M100–S20 (2018).
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection 18 (2012): 268–81.
- Lee K, Chong Y, Shin HB, et al. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobactet species. Clinical Microbiology and Infection 7 (2001): 88–91.
- Bora A, Sanjana R, Jha BK, et al. Incidence of metallo-beta-lactamase producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in central Nepal. BMC Res Notes 7 (2014): 557.
- Franklin C, Liolios L, Peleg AY. Phenotypic Detection of Carbapenem-Susceptible Metallo-β-Lactamase-Producing Gram-Negative Bacilli in the Clinical Laboratory. J Clin Microbiol 44 (2006): 3139–44.
- Bratu S, Landman D, Alam M, et al. Detection of KPC Carbapenem-Hydrolyzing Enzymes in Enterobacter spp. from Brooklyn, New York. Antimicrob Agents Chemother 49 (2005): 776–8.
- Mendes RE, Kiyota KA, Monteiro J, et al. Rapid Detection and Identification of Metallo-β-Lactamase-Encoding Genes by Multiplex Real-Time PCR Assay and Melt Curve Analysis. J Clin Microbiol 45 (2007): 544–7.
- Debnath J, Das PK. Debnath J, et al, Bacteriological profile and antibiotic susceptibility pattern of neonatal septicemia in a tertiary care hospital of Tripura. Indian J Microbiol Res 2 (2015): 238-243,.
- Chakraborty S, Mohsina K, Sarker PK, et al. Prevalence, antibiotic susceptibility profiles and ESBL production in Klebsiella pneumoniae and Klebsiella oxytoca among hospitalized patients. Period Biol 23 (2016): 118.
- Akter J, Azad Chowd AMM, MAF. Study on Prevalence and Antibiotic Resistance Pattern of Klebsiella Isolated from Clinical Samples in Southeast Region of Bangladesh. American J of Drug Discovery and Development 4 (2013): 73–9.
- Nirwati H, Sinanjung K, Fahrunissa F, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc 13 (2019): 20.
- Aljanaby AAJ, Alhasani AHA. Virulence factors and antibiotic susceptibility patterns of multidrug resistance Klebsiella pneumoniae isolated from different clinical infections. Afr J Microbiol Res 10 (2016): 829–843.
- Khatun MM, Sharif MM, Haque AE, et al. Bacteriological Profile of ESBL Producing Bacteria With Their Antibiotic Resistance Pattern. KYAMC j 10 (2019): 7–12.
- Ahmed I, Rabbi MdB, Sultana S. Antibiotic resistance in Bangladesh: A systematic review. International Journal of Infectious Diseases 80 (2019): 54–61.
- Kawser Z, Shamsuzzaman SM. Association of Virulence with Antimicrobial Resistance among Klebsiella pneumoniae Isolated from Hospital Settings in Bangladesh. International Journal of Applied & Basic Medical Research 12 (2022): 123–9.
- Hasan J, Hosen S, Bachar SC. The resistance growing trend of common gramnegative bacteria to the potential antibiotics over three consecutive years: a single center experience in Bangladesh. PPIJ 25 (2019): 7(3).
- Chakraborty S. Prevalence, antibiotic susceptibility profiles and ESBL production in Klebsiella pneumoniae and Klebsiella oxytoca among hospitalized patients. Period Biol 118 (2016): 53–8.
- Saba Riaz. Prevalence and comparison of Beta-lactamase producing Escherichia coli and Klebsiella spp from clinical and environmental sources in Lahore, Pakistan. Afr J Microbiol Res 6 (2012).
- Farzana R, Shamsuzzaman S, Mamun KZ. Isolation and molecular characterization of New Delhi metallo-beta-lactamase-1 producing superbug in Bangladesh. J Infect Dev Ctries 7 (2013): 161–8.
- Eftekhar F, Naseh Z. Extended-spectrum β-lactamase and carbapenemase production among burn and non-burn clinical isolates of Klebsiella pneumoniae. Iran J Microbiol 7 (2015): 144–9.
- Metwally L, Gomaa N, Attallah M, et al. High prevalence of Klebsiella pneumoniae carbapenemase-mediated resistance in K. pneumoniae isolates from Egypt. East Mediterr Health J 19 (2013): 947–52.
- Nordmann P, Poirel L, Dortet L. Rapid Detection of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18 (2012): 1503–7.
- Farzana R, Shamsuzzaman S, Mamun KZ. Isolation and molecular characterization of New Delhi metallo-beta-lactamase-1 producing superbug in Bangladesh. J Infect Dev Ctries 7 (2013): 161–8.
- Rahman M, Shukla SK, Prasad KN, et al. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. International Journal of Antimicrobial Agents 44 (2014): 30–7.
- Queenan AM, Bush K. Carbapenemases: The Versatile β-Lactamases. Clin Microbiol Rev 20 (2007): 440–58.
- Satter S, Shamsuzzaman S. Phenotypic and Genotypic Characterization of Extended-spectrum beta-lactamase Producing Escherichia coli and Klebsiella species Isolated from a Tertiary Care Hospital in Bangladesh. Bangladesh J Med Microbiol 10 (2016): 4–8.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
